Katharina Schiessl
Career Development Fellow
Sainsbury Laboratory Cambridge
University of Cambridge
Bateman Street
Cambridge CB2 1LR
Email: katharina.schiessl@slcu.cam.ac.uk
Connect: @kathschiessl | LinkedIn | ResearchGate
Research Interests
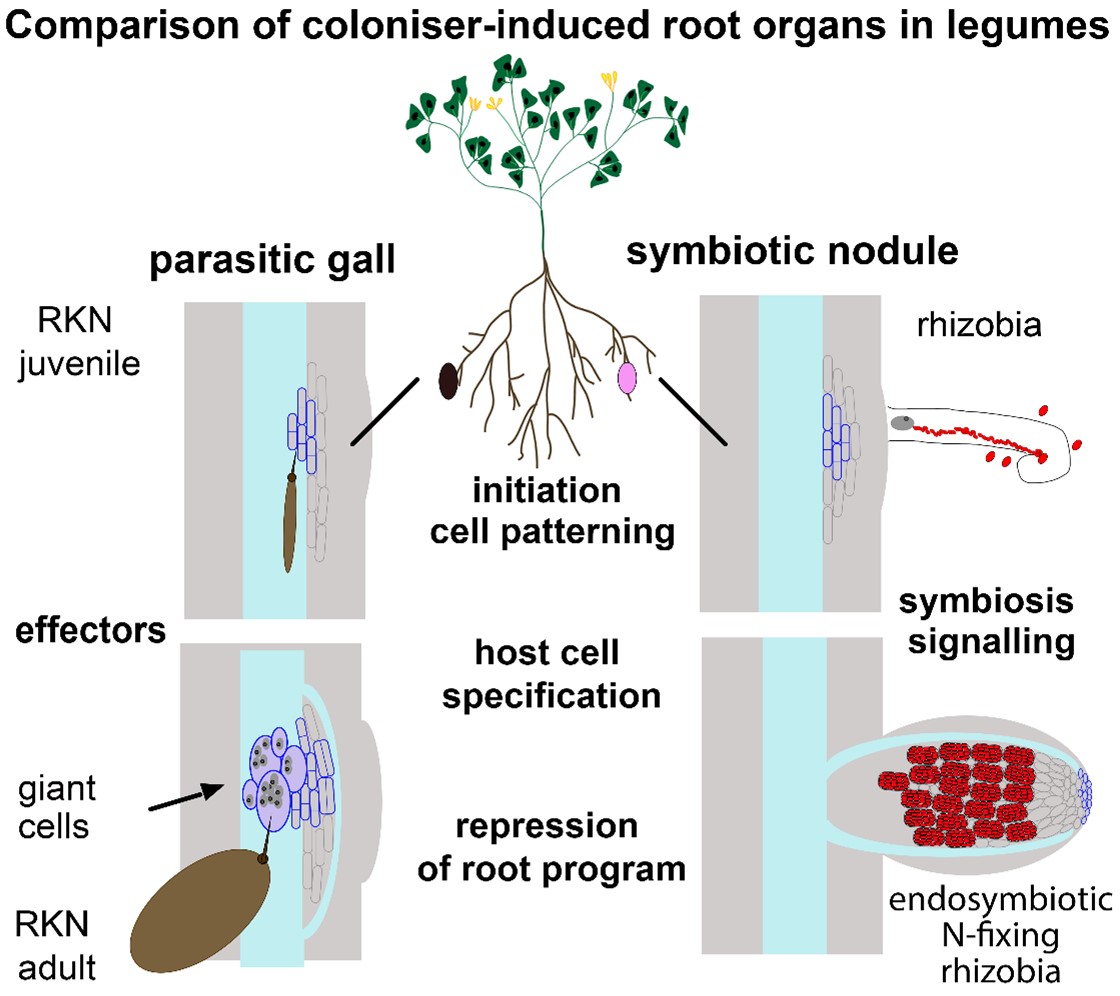
The Schiessl Group is interested in the question of how novel plant organs of diverse form and function can be generated. We use coloniser-induced plant organs such as symbiotic root nodules and parasitic root galls to advance and refine our understanding of the common principles that underpin plant organ diversification.
Unlike animals, plants can grow new organs such as leaves and roots throughout their lives. Some plant colonisers have evolved means to exploit this unique ability of plants for their own purposes, directing the formation of custom-built and sometimes very intricate plant structures referred as galls. Hence these colonisers can be considered as generators of plant organ diversity.
We predominantly use the legume Medicago truncatula (barrel medic), a model for temperate legume crops such as pea, because it is a common host for nodule-inducing nitrogen-fixing bacteria and root-gall-inducing parasitic root-knot nematodes. This provides us with a framework to compare the plant developmental regulatory pathways oflateral roots, symbiotic root nodules and parasitic root galls within the same genetic background and tissue context.
Our aim is to
- genetically dissect and characterize the pre-existing gene regulatory pathways and molecular processes in plants that are recruited and modified for the development of parasitic root galls and symbiotic root nodules;
- genetically dissect and functionally characterise the molecular processes that differentiate host cells into endosymbiotic cells in nodules and giant feeding site cells in parasitic galls;
- to identify and characterise the molecular means by which these colonisers access and modify these plant developmental pathways;
For this, we use developmental genetics combined with quantitative deep tissue imaging to generate data on gene expression, cell growth, proliferation, and differentiation in coloniser-induced plant organs. Complimentarily, we use genome-scale profiling approaches of gene expression and cis regulatory elements to systematically discover novel components and interactions within gene regulatory networks recruited and modified by the colonisers. Our experimental data build the foundation for computational models explaining the morphogenesis and evolution of these novel and unique organs. Plant galls provide shelter and feeding sites for the colonisers. Therefore, it is not surprising that some gall-inducing organisms are major crop pests. This includes root-knot nematodes that induce root galls in many crop species and are therefore a severe threat to global food security. By contrast, beneficial root nodule-inducing bacteria provide plants with nitrogen, a macronutrient essential for plant growth.
We anticipate that the outcomes of our research will reveal the regulatory networks and cellular processes directing coloniser-induced organ development. Thereby our research brings an “organ-centred”, developmental perspective to these plant-microbe interactions and has the potential to provide future strategies to improve the resilience of our crops against parasitic colonisers and to promote biological nitrogen fixation.
Our Research Objectives
Objective 1: Dissect & characterise the regulatory pathways and cellular processes of coloniser-induced organogenesis via developmental genetics
Our work and works by others have shown that lateral roots, symbiotic nodules and parasitic galls emerge from a conserved organ initiation programme. This prompts the question of what pre-existing endogenous plant developmental pathways are subsequently recruited and modified to differentiate coloniser-induced organs from endogenous organs in form and function.
In particular, plant cells in coloniser-induced organs acquire customised features and functions to serve the specific needs of their colonisers. In nodules, host cells take up rhizobial bacteria intracellularly in membrane-bound vesicles, the symbiosomes, and provide the environment for biological nitrogen fixation. In parasitic galls, root knot nematodes induce giant cells that serve as the sole feeding sites for the females. Despite very different purposes, both host cell types share common features, including an increase in size and nuclear content. Cell type specification in plants is mediated by the coordinated activity of organ identity genes, growth regulators and hormone signalling. We will use developmental genetics to understand how the regulatory pathways investigated in objective 1 impact and modify cellular processes to result in the highly customized features and functions found in endosymbiotic host cells and parasitic giant cells. Given their novel and specialised features, unravelling the cellular mechanisms underlying their differentiation poses an exceptionally intriguing question.
Using genome-scale and developmental genetic approaches, we are characterising the regulatory pathways and cellular processes that underpin the initiation and differentiation of root-related organs associated with the accommodation of beneficial nitrogen-fixing bacteria and parasitic root knot nematodes.
Objective 2: Identify & characterise the means by which colonisers direct plant organ development
Post-embryogenic organ development in plants is generally triggered in response to external signals such as light or nutrients. Similarly, parasitic and symbiotic colonisers both rely on secreted signals to initiate and direct the development of custom-built plant organs.
Beneficial rhizobia and legume hosts communicate via bacterial nodulation factors and the plant symbiosis signalling pathway throughout the colonisation process. However, it remains unclear how the epidermal infection process is coordinated with the initiation and development of the nodule primordium in the inner tissue layers right below the site of infection.
In particular, the signal that crosses tissue layers to temporally and spatially coordinate these two processes has yet to be discovered. Recent work characterized a set of transcriptional regulators with restricted expression in nodule primordium cells that are not only required for early nodule development but also for the progression of bacterial infection in the outer tissue layers. This provides a promising framework for the discovery of the elusive signal that coordinates the progression of the bacterial infection process with the development of the nodule primordium in the inner tissue layers.
Some small secreted peptides (effectors) encoded in the genomes of plant parasitic nematodes can mimic small, secreted plant peptides with functions in plant organ development, providing promising targets for root knot nematode interference. Discovering effectors that specifically interfere with developmental targets in the host helps to further explore how plant developmental plasticity can be exploited beyond endogenous constraints, exemplified by the unique features of the giant cells. For this, we aim to establish and foster collaborations with groups working on molecular plant-microbe interactions and effector biology.
Research published in the media and science features
- Overlap in lateral root and nodule development brings self-fertilising cereals one step closer | Sainsbury Laboratory
- Growing excitement about self‑fertilising crops | The Times
Key Publications
Tak Lee, Martina Orvosova, Morgane Batzenschlager, Marcelo Bueno Batista, Paul C. Bailey, Nadia A. Mohd-Radzman, Aram Gurzadyan, Naomi Stuer, Kirankumar S. Mysore, Jiangqi Wen, Thomas Ott, Giles E.D. Oldroyd, Katharina Schiessl (2024) Light-sensitive short hypocotyl genes confer symbiotic nodule identity in the legume Medicago truncatula. Current Biology. https://doi.org/10.1016/j.cub.2024.01.018
Colleen Drapek, Nadiatul Radzman-Mohd, Annalisa Rizza, Katharina Schiessl, Fabio Dos Santos Barbosa, Jiangqi Wen, Giles E.D. Oldroyd, Alexander M. Jones: Cellular gibberellin dynamics govern indeterminate nodule development, morphology and function. bioRxiv 2023 (https://doi.org/10.1101/2023.09.09.556959) PRE.
Schiessl K†, Lee T, Orvosova M, Bueno-Batista M, Stuer N, Bailey P C, Mysore K S, Wen J and Oldroyd G E D: Light sensitive short hypocotyl (LSH) confer symbiotic nodule identity in the legume Medicago truncatula. bioRxiv 2023 (https://www.biorxiv.org/content/10.1101/2023.02.12.528179v1) PRE.
Roy S, Jerez T, Zhang S, Liu W, Schiessl K, Boschiero C, Lee H-K, Zhao PX, Murray JD, Oldroyd GED, Scheible WR, Udvardi M: The peptide GOLVEN10 controls nodule and lateral root organogenesis and positioning along the longitudinal root axis. bioRxiv 2022 (https://www.biorxiv.org/content/10.1101/2022.05.06.490929v1) PRE.
Feng, J, Lee T, Schiessl K, and Oldroyd G E D: Processing of NODULE INCEPTION controls transition to nitrogen fixation in root nodules. Science 2021 (DOI: 10.1126/science.abg2804).
D'Ario M, Tavares R, Schiessl K, Desvoyes B, Gutierrez C., Howard M and Sablowski R. Cell size controlled in plants using DNA content as an internal scale. Science 2021 ( DOI: 10.1126/science.abb4348).
Yang W, Cortijo S, Korsbo N, Roszak P, Schiessl K, Gurzadyan A, Wightman R, Jönsson H, Meyerowitz E. Molecular mechanism of cytokinin-activated cell division in Arabidopsis. Science 2021 (DOI: 10.1126/science.abe2305).
Schiessl K, Lilley J L S, Lee T, Tamvakis I, Kohlen W, Bailey P C, Thomas A, Luptak J, Ramakrishnan K, Carpenter M D, Mysore K S, Wen J, Ahnert S, Grieneisen V A and Oldroyd G E D: NODULE INCEPTION recruits the lateral root developmental program for symbiotic nodule organogenesis in Medicago truncatula. Current Biology 2019 (DOI: 10.1016/j.cub.2019.09.005).
Magne K, Couzigou J-M, Schiessl K., Liu S, George J, Zhukov V, Sahl L, Boyer F, Iantcheva A, Mysore K S, Wen J, Citerne S, Oldroyd G E D, Ratet P: MtNODULE ROOT1 and MtNODULE ROOT2 Are Essential for Indeterminate Nodule Identity. Plant Physiology 2018 (DOI: 10.1104/pp.18.00610).
Mislata A, Bencivenga S, Bush M, Schiessl K, Boden S and Sablowski R: DELLA genes restrict inflorescence meristem function independently of plant height. Nature Plants 2017 (DOI: 10.1038/s41477-017-0003-y).
Mislata A, Schiessl K, Sablowski R: Active control of cell size generates spatial detail during plant organogenesis. Current Biology 2015 (DOI:10.1016/j.cub.2015.10.008).
Schiessl K, Muino J M, Sablowski R: Arabidopsis JAGGED links floral organ patterning to tissue growth by repressing Kip-related cell cycle inhibitors. PNAS 2014 (DOI:10.1073/pnas.1320457111).
Sauret-Güeto S, Schiessl K, Bangham A, Sablowski R, Coen E: JAGGED Controls Arabidopsis Petal Growth and Shape by Interacting with a Divergent Polarity Field. PLOS Biology 2013 (DOI: 10.1371/journal.pbio.1001550).
Schiessl K, Kausika S, Southam P, Bush M, Sablowski R: JAGGED controls growth anisotropy and coordination between cell size and cell cycle during plant organogenesis. Current Biology 2012 (DOI: 10.1016/j.cub.2012.07.020