Dr François Nédélec
Research Group Leader
Sainsbury Laboratory Cambridge
University of Cambridge
47 Bateman Street
Cambridge CB2 1LR
Office Phone: 01223 761159
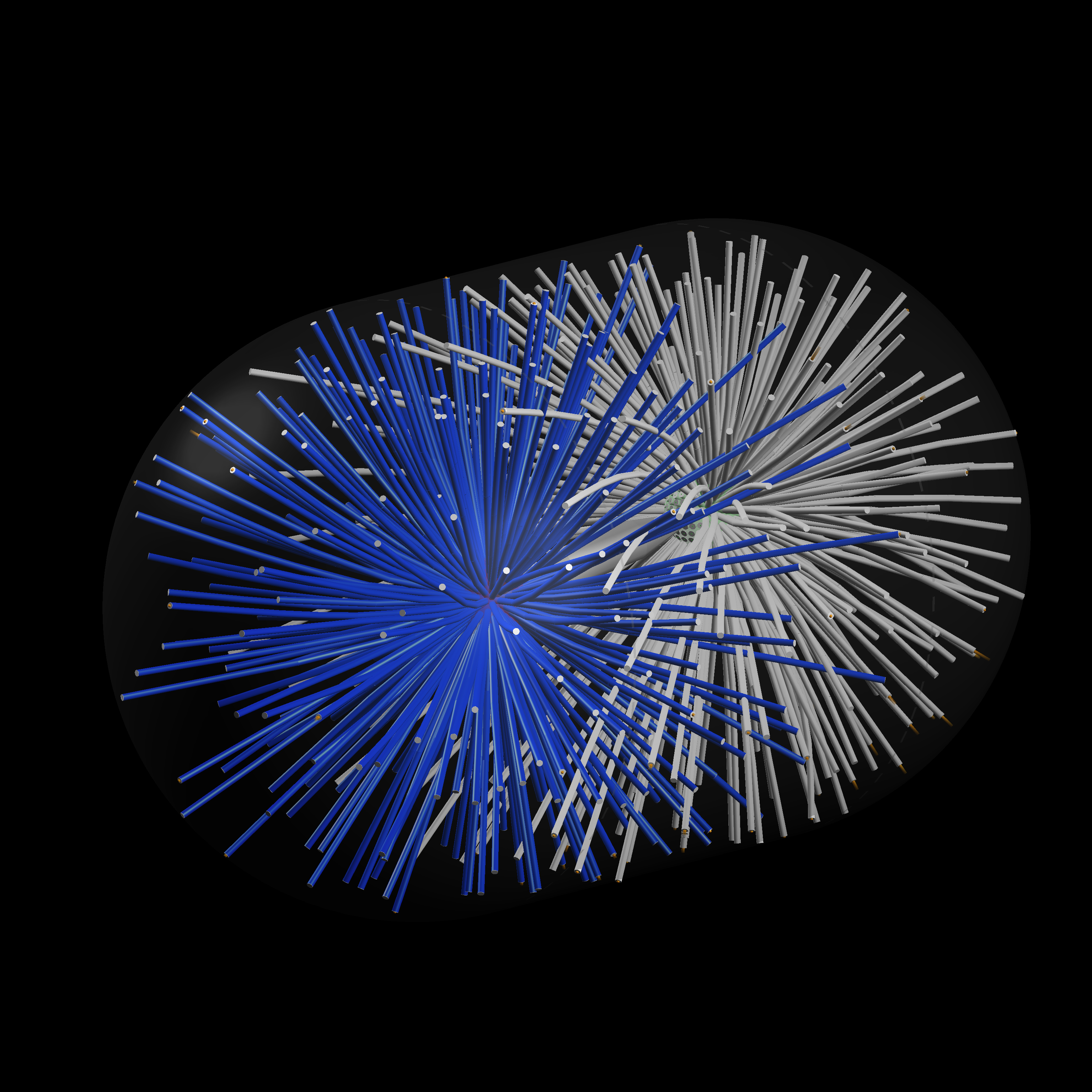
A Model of the Assembly of Mitotic Spindle found in Roots of Arabidopsis.
Research Interests
Dr Nédélec is growing and facilitating a user community to use cytosim – a generic platform to model the cytoskeleton. Cytosim is an Open Source simulation platform that can simulate systems composed of filaments with bending flexibility, achieving a high level of realism in 2D or 3D. It can simulate the interactions of thousands of filaments with millions of associated proteins such as molecular motors according to the laws of mechanics and stochastic reaction-kinetics. Cytosim has been applied to various problems involving actin or microtubules networks, and has also been used to guide the development of analytical theory. The filaments can represent microtubules, F-actin or any other type of filament. Cytosim can simulate systems that are mechanically percolated using a custom Brownian Dynamics engine in which chemical reactions are modeled stochastically. It can simulate large systems composed of tens of thousands of filaments rapidly enough to enable their biophysical analysis. Cytosim has been validated in many studies, relating to processes ranging from nuclear positioning, spindle assembly and actin network organisation.
Mechanistic Analysis of Mitotic Spindle Assembly
Mitotic spindles are large cellular structures made for segregating the chromosomes during eukaryotic cell division. This function is universal, and essential for sustained proliferation of cells. Dr Nédélec is using experimental and theoretical methods to analyse the organisation of spindles in plants and yeast.
Cell Morphogenesis
Dr Nédélec is particularly interested in exploring how the shape of plant cells are determined and how the cell wall and cytoskeleton are interacting to modulate the cell shape. A key question is to explore why particular cell shapes are optimal, with an emphasis on how the shape confers advantageous mechanical properties.
Selected Publications
Determinants of Polar versus Nematic Organization in Networks of Dynamic Microtubules and Mitotic Motors. Roostalu J, Rickman J, Thomas C, Nédélec F, Surrey T. Cell 175(3):796-808.e14. doi: 10.1016/j.cell.2018.09.029 (2018)
Systematic Nanoscale Analysis of Endocytosis Links Efficient Vesicle Formation to Patterned Actin Nucleation. Mund M, van der Beek JA, Deschamps J, Dmitrieff S, Hoess P, Monster JL, Picco A, Nédélec F, Kaksonen M, Ries J. Cell 174(4) doi: 10.1016/j.cell.2018.06.032 (2018)
A theory that predicts behaviors of disordered cytoskeletal networks J. Belmonte, M. Leptin and F. Nédélec. Molecular Systems Biology, 10.15252/msb.20177796 (2017)
Asymmetric division of contractile domains couples cell positioning and fate specification
Maître JL, Turlier H, Illukkumbura R, Eismann B, Niwayama R, Nédélec F, Hiiragi T.
Nature 536(7616):344-348. doi: 10.1038/nature18958 (2016)
Mechanical design principles of a mitotic spindle J. Ward, H. Roque, C. Antony and F. Nédélec eLife; 10.7554/eLife.03398 (2014)
Asymmetric division of contractile domains couples cell positioning and fate specification
J-L. Maitre, H. Turlier, R. Illukkumbura, B. Eismann, R. Niwayama, F. Nédélec and T. Hiiragi.
Nature 536, 344-348 (2016)
Pulsatile cell-autonomous contractility drives compaction in the mouse embryo
J-L. Maitre, R. Niwayama, H. Turlier, F. Nédélec and T. Hiiragi
Nature Cell Biology, 17, 849—55 (2015)
Patterns of molecular motors that guide and sort filaments
B. Rupp and F. Nédélec
Lab on a chip, 12, 4903—10 (2012)
Katanin contributes to interspecies spindle length scaling in Xenopus
R. Loughlin, J. Wilbur, McNally F, F. Nédélec*, and R. Heald*. *Corresponding authors.
Cell 147:1397—407 (2011)
A computational model predicts Xenopus meiotic spindle organization
R. Loughlin,
R. Heald* and F. Nédélec*. *Corresponding authors
Journal of Cell Biology 191(7):1239—49 (2010)
Chromatin shapes the mitotic spindle
Dinarina, C. Pugieux, M. Mora-Corral, M. Loose, J. Spatz, E. Karsenti and F. Nédélec
Cell 138:502—13 (2009)
Cortical microtubule contacts position the spindle in C.elegans embryos
C. Kozlowski, M. Srayko and F. Nédélec
Cell 129 (3):499—510 (2007)
Crosslinkers and motors organize dynamic microtubules to form stable bipolar arrays in fission yeast
M. Janson, R. Loughlin, I. Loiodice, C. Fu, D. Brunner, F. Nédélec* and P. Tran*. *Corresponding authors.
Cell 128:357—68 (2007)
Self-organization of microtubules and motors
F. Nédélec, T. Surrey, A.C. Maggs, S. Leibler
Nature 389:305—8 (1997)

