James Locke
Professor of Quantitative Plant Development
Research Group Leader
Royal Society University Research Fellow
University of Cambridge
47 Bateman Street
Cambridge CB2 1LR
Email: james.locke@slcu.cam.ac.uk
Office Phone: 01223 761110
Research Interests
In order to gain a quantitative understanding of the gene circuits that underlie plant development, it is critical to examine gene regulation at the single-cell level through time. Single-cell analysis has revealed that gene regulation can be surprisingly dynamic and heterogenous, in ways that are not identified by traditional approaches that take an average from a population. In our lab we use a combination of single-cell time-lapse microscopy, mathematical modelling and synthetic biology approaches to understand the design principles of dynamic gene regulation. We use Arabidopsis as a model system for understanding the role of dynamic gene regulation in development, whilst also testing our conceptual ideas in simpler systems such as Cyanobacteria and B. subtilis. In Arabidopsis we are building on our work in these simpler systems to reveal the functional role of dynamic, and even stochastic, gene regulation in development.
Selected Recent Publications
CP Schwall, TE Loman, BMC Martins, S Cortijo, C Villava, V Kusmartsev, T Livesey, T Saez, JCW Locke, Tunable phenotypic variability through an autoregulatory alternative sigma factor circuit, Molecular Systems Biology, 2021; 17:e9832
K Abley, P Formosa-Jordan, H Tavares, EYT Chan, M Afsharinafar, O Leyser^, JCW Locke^, An ABA-GA bistable switch can account for natural variation in the variability of Arabidopsis seed germination time , eLife, 2021; 10:e59485
E Nadezhdin, N Murphy, N Dalchau, A Phillips, JCW Locke, Stochastic pulsing of gene expression enables the generation of spatial patterns in Bacillus subtilis biofilms, Nature communications, 2020; 11 (1), 1-12
S Cortijo, M Bhattarai, JCW Locke^, SE Ahnert^, Co-expression networks from gene expression variability between genetically identical seedlings can reveal novel regulatory relationships, Frontiers in plant science, 2020; 11:599464
M Greenwood, M Domijan, PD Gould, AJW Hall, JCW Locke, Coordinated circadian timing through the integration of local inputs in Arabidopsis thaliana, PLoS biology, 2019; 17 (8), e3000407
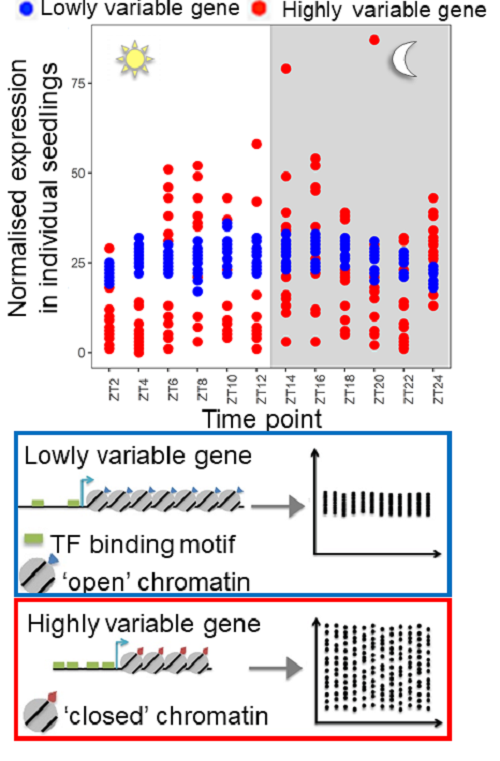
S Cortijo, Z Aydin, S Ahnert, JCW Locke, Widespread inter-individual gene expression variability in Arabidopsis thaliana , Molecular systems biology, 2019; 15 (1), e8591
O Patange, C Schwall, M Jones, C Villava, DA Griffith, A Phillips, JCW Locke, Escherichia coli can survive stress by noisy growth modulation, Nature communications, 2018; 9 (1), 1-11
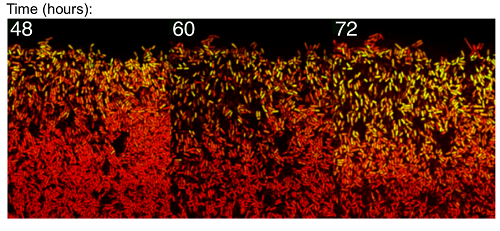
BMC Martins, AK Tooke, P Thomas, JCW Locke, Cell size control driven by the circadian clock and environment in cyanobacteria, PNAS, 2018; (48), E11415-E11424
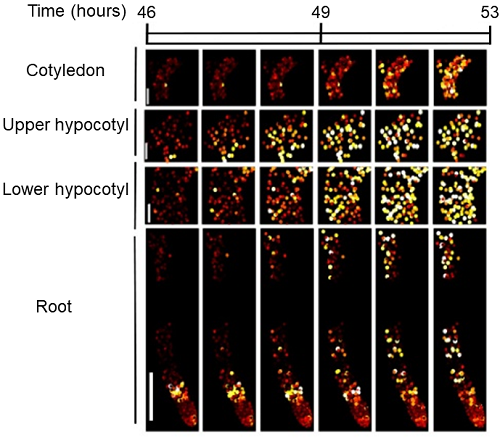
P Gould*, M Domijan*, M Greenwood, IT Tokuda, H Rees, L Kozma-Bognar, AJW Hall^, JCW Locke^, Coordination of robust single cell rhythms in the Arabidopsis circadian clock via spatial waves of gene expression, eLife, 2018; 7, e31700
J Park, M Dies, Y Lin, S Hormoz, SE Smith-Unna, S Quinodoz, M Hernández-Jiménez, J Garcia-Ojalvo^, JCW Locke^, MB Elowitz^, Molecular time-sharing through dynamic pulsing in single cells, Cell Systems, 2018; 6 (2), 216-229.
B Landrein, P Jordan-Formosa, C Melnyk, C Schuster, W Yang, C Turnbull, EM Meyerowitz^, JCW Locke^, H Jönsson^, Nitrate modulates stem cell dynamics by regulating WUSCHEL expression through cytokinins, PNAS, 2018; Feb 6;115(6):1382-1387.
HM Meyer*, J Teles*, P Formosa-Jordan*, Y Refahi, R San-Bento, G Ingram, H Jönsson^, JCW Locke^, AHK Roeder^, Fluctuations of the transcription factor ATML1 generate the pattern of giant cells in the Arabidopsis sepal, eLife, 2017; e19131.
BMC Martins*, AK Das*, L Antunes, JCW Locke, Frequency doubling in the cyanobacterial circadian clock, Molecular Systems Biology, 2016; 12 (12), 896. https://www.embopress.org/doi/full/10.15252/msb.20209832
Selected Recent Reviews
CN Micklem, JCW Locke, Cut the noise or couple up: Coordinating circadian and synthetic clocks, iScience, 2021; 24 (9), 103051
S Cortijo, JCW Locke, Does gene expression noise play a functional role in plants? Trends in Plant Science, 2020; 25 (10), 1041-1051
M Greenwood, JCW Locke, The circadian clock coordinates plant development through specificity at the tissue and cellular level, Current opinion in plant biology, 2020; 53, 65-72
K Abley, JCW Locke, HMO Leyser, Developmental mechanisms underlying variable, invariant and plastic phenotypes, Annals of Botany, 2016; 117 (5), 733-748.
BMC Martins, JCW Locke, Microbial individuality: how single-cell heterogeneity enables population level strategies, Current Opinion in Microbiology, 2015; 24, 104-112.
*Joint first authors
^Joint corresponding authors