Open source platforms
We have developed a number of public open-source software for bioinformatics. imaging processing and modelling applications.
Bioinformatics
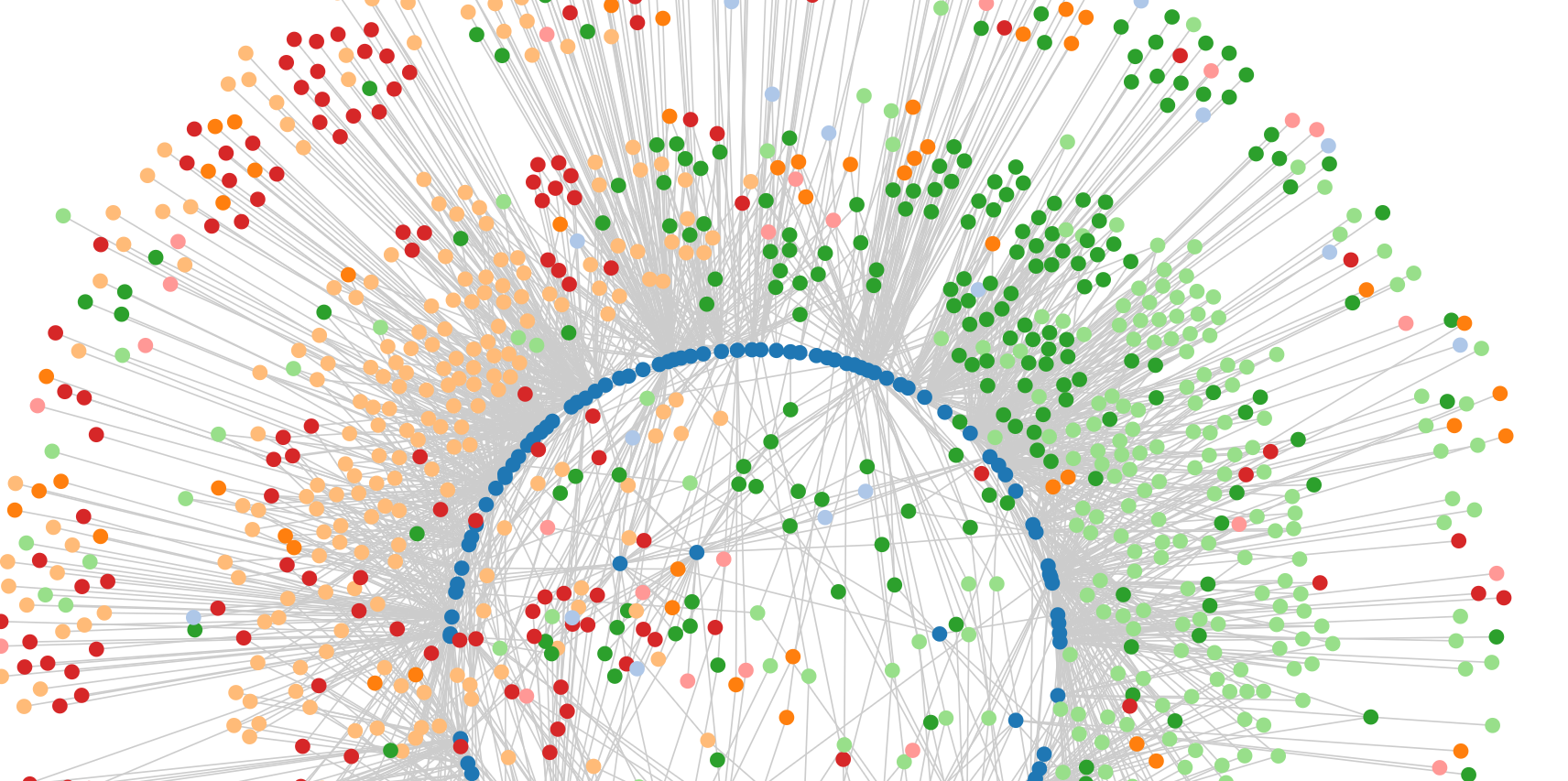
Ara-BOX-cis
A visualisation of the G-box regulatory network in Arabidopsis
This tool provides ways of visualising the network of bHLHs/bZIPs and their predicted regulatory targets among genes that are expressed in seedlings and that contain perfect G-boxes (CACGTG) within 500bp upstream of their TSS. A more detailed description of how this network was generated can be found in the publication The G-Box Transcriptional Regulatory Code in Arabidopsis (Plant Physiology 2017) .
Visit the website at araboxcic.org
SecretSanta
Open-source software tool that automates the rapid and accurate prediction of secretomes
The SecretSanta package provides an R interface for the integrative prediction of extracellular proteins that are secreted via classical pathways. Automated pipelines created within the SecretSanta framework can be applied to large protein sets to facilitate comparison of secretomes across multiple species or under various conditions. Source code, installation instructions, documentation and use case scenarios are available at https://github.com/gogleva/SecretSanta.
Read this article on the development and use of SecretSanta.
Image Processing
Costanza image segmentation software
Image segmentation software implemented as an ImageJ plugin (Java) and batch mode version developed by the Jönsson group. It can be used for 2D membrane segmentation (e.g. Jönsson et al (2006), PNAS) and 3D nuclei segmentation (e.g. Meyer et al (2017), eLife. More information and src code at Costanza.
Modelling
Cytosim cytoskeleton simulation
Cytosim is a cytoskeleton simulation designed to handle large systems of flexible filaments with associated proteins such as molecular motors. It is a versatile base that has been used to study actin and microtubule systems in 1D, 2D and 3D. It is built around a core cross-platform C++ engine. It runs on UNIX, Mac OSX, GNU/Linux and within Cygwin on Windows. The code is modular and extensible, making Cytosim a convenient base that can be customized to meet particular tasks.
More information can be found on the Cytosim Gitlab page.
Organism
The Organism-Tissue Simulator is a C++ software for simulating biological systems developed by the Jönsson group. It has been used to simulate plant tissues, bacteria colonies and budding yeast colonies. More information can be found on the gitLab page.
