Internal leaf patterning
The Whitewoods Group aims to understand how plants pattern the inside of their leaves.
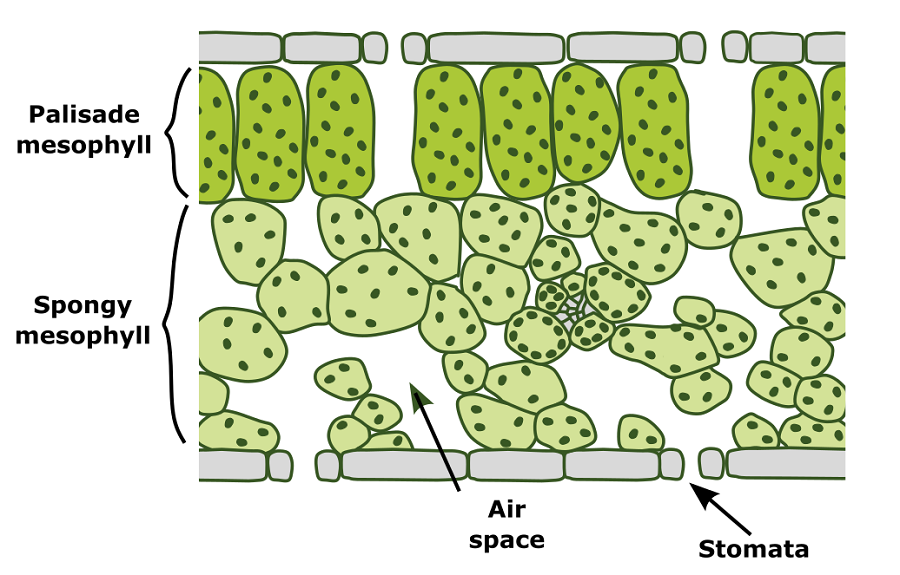
From the surface leaves look like flat green sheets, but inside is a hidden world of complexity: many leaves contain two primary photosynthetic cell types surrounded by intercellular air spaces. The upper half contains densely packed palisade mesophyll cells with only small air spaces, whereas the lower half contains irregularly shaped spongy mesophyll cells surrounded by larger air spaces. This arrangement is thought to maximise light harvesting in the top half and gas exchange and light scattering in the lower half to increase overall photosynthetic efficiency and minimise water loss.
Our research aims to answer two main questions:
- How are intercellular air spaces patterned?
- What specifies palisade and spongy mesophyll cell fate?
To answer these questions we combine computational modelling with genetic and developmental analysis in the flat-leaved model plant Arabidopsis thaliana and the floating aquatic carnivorous plant Utricularia gibba, among others. We have several projects that feed into each other to give an integrated understanding of internal leaf patterning:
Diagram of a leaf cross section showing location of palisade and spongy mesophyll cells with surrounding intercellular air spaces. Graphic by Chris Whitewoods.
Genetic basis of air space development and evolution
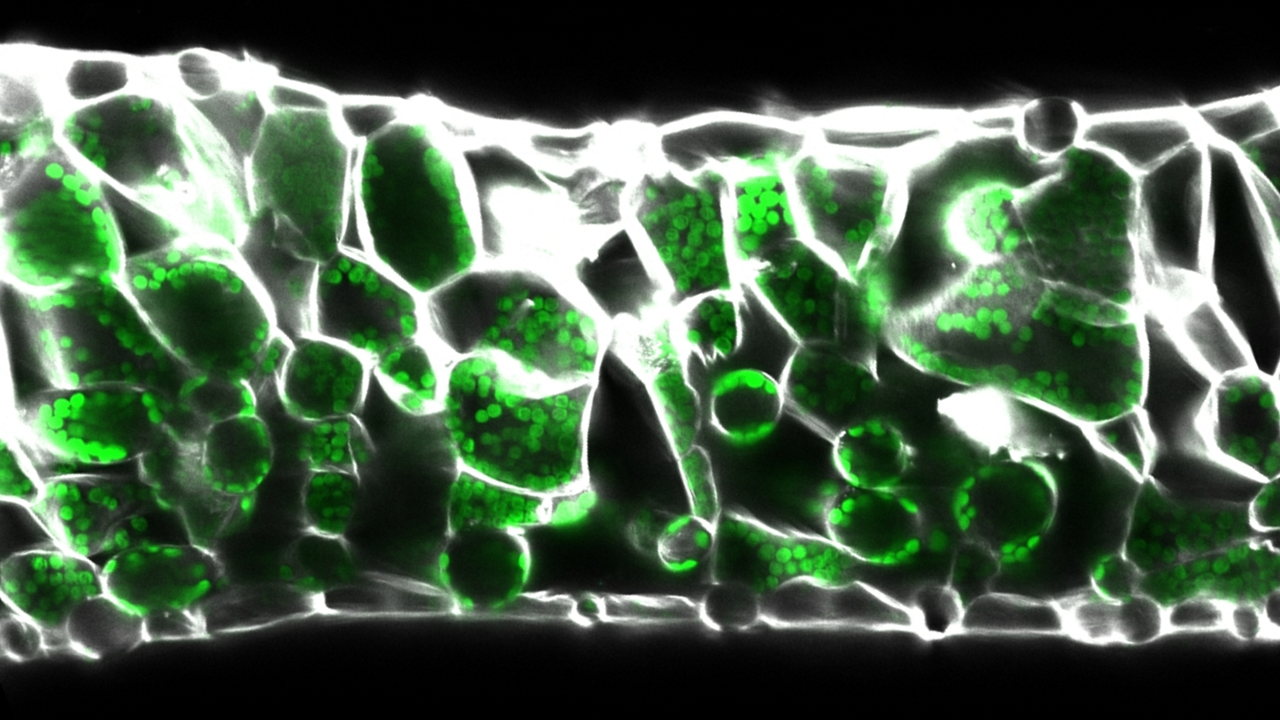
Intercellular air spaces make up to 70% of leaf volume and are vital for leaf function. In flat leaves air spaces are associated with stomata and maximise gas exchange for photosynthesis, while in aquatic plants these air spaces have become enlarged and aid floatation. However, despite being a major part of leaf structure we do not understand how air spaces form, or how new air space patterns evolve.
To identify novel regulators of air space formation we perform forward and reverse genetic screens in A. thaliana and U. gibba. We identify plants with altered air spaces, quantify differences in patterning and map the genes that underlie these phenotypes. We combine these genetic approaches with computational modelling and inducible genetic changes to understand exactly how these genes influence development and control air space formation. We also perform comparative experiments to investigate how these genes have been modified through evolution to control differences in air space patterning between plant species.
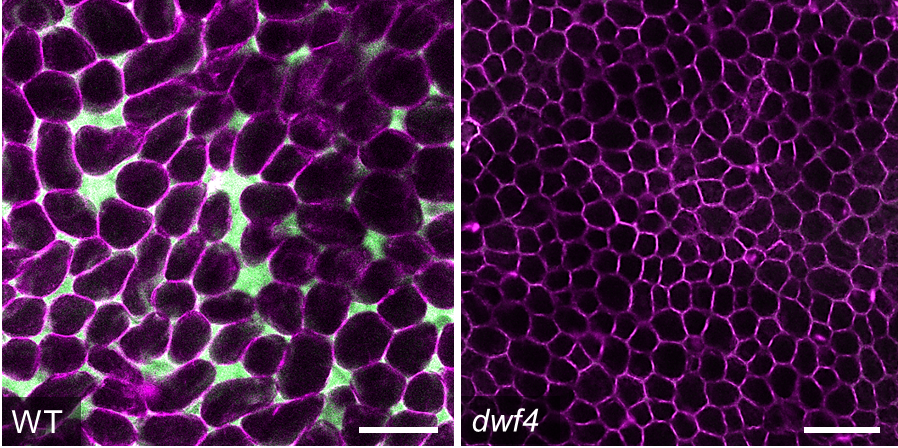
Palisade mesophyll cells of a wild type A. thaliana leaf (left) and the dwf4 mutant with reduced air spaces (right). Samples are stained with Nile Red in Silicon oil to visualise air spaces (in green) and cell membranes (in magenta). Images by James Fitzsimons.
Cell division, growth and intercellular adhesion in air space development
Leaf air spaces form by initially connected cells separating, but how plants regulate this process is unknown. We generate computational models to predict the effect of cell division, expansion and adhesion on air space formation, and test these hypotheses using inducible genetic changes to alter growth, cell wall properties and intercellular adhesion. These approaches allow us to investigate how differential growth and cell adhesion contribute to air space formation.
Identifying regulators of Spongy and Palisade cell fate
To identify genes that control mesophyll cell fate we are undertaking a mutant screen to find plants that lack either spongy or palisade cell fate (see here for more information). Once we have identified regulatory genes we will test how they work to control development, how they respond to the environment and how they interact with other known regulators of leaf development to pattern the leaf in three dimensions.
We are also investigating how chloroplasts control internal leaf patterning. Many classic mutants with defective chloroplasts have an increased proportion of intercellular air space and more irregular mesophyll cell shapes. We are performing a range of experiments to understand whether chloroplasts control spongy vs. palisade mesophyll cell fate, regulate mesophyll morphogenesis or simply promote cell death when they are defective.
Key publications
Kelly-Bellow R, Lee K, Kennaway R, Barclay JE, Whibley A, Bushell C, Spooner J, Yu M, Brett P, Kular B, Cheng S, Chu J, Xu T, Lane B, Fitzsimons J, Xue Y, Smith RS, Whitewoods CD, Coen E. 2023. Brassinosteroid coordinates cell layer interactions in plants via cell wall and tissue mechanics. Science. 2023. doi: 10.1126/science.adf0752. https://doi.org/10.1126/science.adf0752
Whitewoods CD*, Gonçalves B*, Cheng J*, Cui M, Kennaway R, Lee K, Bushell C, Yu M, Piao C. and Coen E. 2020. Evolution of carnivorous traps from planar leaves through simple shifts in gene expression. Science. https://doi.org/10.1126/science.aay5433
Lee K*, Bushell C*, Kiode Y*, Fozard J, Piao C, Yu M, Newman J, Whitewoods CD, Avondo J, Kennaway R, Maree A, Cui M. and Coen E. 2019. Shaping of a three-dimensional carnivorous trap through modulation of a planar growth mechanism. PLOS Biology. https://doi.org/10.1371/journal.pbio.3000427
Dennis R, Whitewoods CD and Harrison CJ. 2019. Quantitative methods for like-for-like comparison in analyses of Physcomitrella patens phyllid development. Journal of Bryology. https://doi.org/10.1080/03736687.2019.1668109
Whitewoods CD*, Cammarata J*, Nemec Venza Z, Sang S, Crook AD, Aoyama T, Wang X, Waller M, Kamisugi Y, Cuming A, Svövényi P, Nimchuk ZL, Roeder AHK, Scanlon MJ and Harrison CJ. 2018. CLAVATA was a genetic novelty for the morphological innovation of 3D growth in land plants. Current Biology. https://doi.org/10.1016/j.cub.2018.05.068
Reviews
Whitewoods CD. Riddled with holes: Understanding air space formation in plant leaves. PLoS Biology. 2021;19(12):e3001475. http://dx.doi.org/10.1371/journal.pbio.3001475
Whitewoods CD. 2020. Utricularia: Quick Guide. Current Biology
Whitewoods CD and Coen E. 2017. Growth and development of three-dimensional plant form. Current Biology 27: R910-R918. https://doi.org/10.1016/j.cub.2017.05.079