Plant biomathematicians at the Sainsbury Laboratory Cambridge University (SLCU) have developed a new tool that enables them to listen in on cell-to-environment conversations. The tool’s wide-ranging application means it could help speed-up the development of strategies to combat plant and animal pathogens.

SLCU plant biomathematicians, Dr Anna Gogleva and Dr Hajk-Georg Drost, have developed a toolset that can be applied to study secretomes in any species across the whole tree of life.
Living cells engage with their environment by secreting different types of proteins, some of which act as communication units in a cell-to-cell molecular language. The total abundance and diversity of the proteins secreted by an organism is called its “secretome” and is unique to each organism. To understand the multitude of interactions between cells of the same organism, or even between different organisms, scientists need to decipher this molecular language.
Secretomes may facilitate beneficial interactions, such as in the symbiotic relationship formed between soil fungi and plant roots or help some pathogenic organisms to trick their hosts into allowing them to invade and proliferate. By “eavesdropping” on cell-to-cell conversations, scientists can generate their own “messages” to disrupt detrimental and to reinforce beneficial interactions. A major challenge in deciphering secretomes, however, is the ability to efficiently identify all the individual proteins contributing to the overall language.
|
Secretomes and plant-microbe interactions Scientists are often interested in secreted proteins (extracellular proteins), which are vital for pathogens colonising a host organism to establish a hospitable environment. Also, the host secretes proteins to protect or defend themselves. Knowing which proteins are secreted by which types of host and microbial cells helps researchers to develop strategies for effective pathogen control.
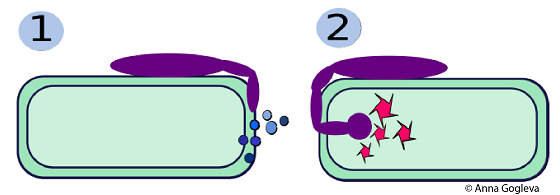
Above diagram: (1) A pathogenic microbe (purple) has latched onto a plant cell and is secreting proteins that destroy the plant cell wall to allow entry. (2) The pathogen secretes a second class of proteins into the plant cell to take-over metabolic control of the cell. Scientists are working to stop pathogens dead in their tracks by developing therapies that target these secretomes. |
Plant Bioinformaticians at SLCU have been working to simplify and speed-up this deciphering process. They have developed an open source software tool that automates the rapid and accurate prediction of secretomes and is optimised to drastically reduce the computation and analytics time from months or weeks down to seconds.
The toolset provided in the package can be applied to study secretomes in any species across the whole tree of life – prokaryotes (bacteria) and eukaryotes (protozoa, algae, fungi, plants and animals).

The package developer, SLCU Research Associate and member of the Schornack research group, Dr Anna Gogleva, came up with the idea for the tool after becoming frustrated by having to repetitively reformat data and writing slightly different modifications of the same pipeline that would be suitable for a number of important plant disease causing pathogens.
“There are numerous programs that have been developed independently over the past 15 years to predict secreted proteins,” Dr Gogleva said. “Each of these tools has been built to have a specific purpose, but researchers often rely on multiple tools to more confidently identify the full set of secreted proteins. While the individual tools are good on their own, they don’t work well together – they are written in different programming languages; they require input data to be formatted differently; some are not capable of processing many protein sequences at a time; they can’t be run in parallel and thus are not scalable; and the results are often incompatible.
“Rather than to reinvent the wheel, I thought that the most effective thing would be to combine these different tools and tool versions into a user-friendly interface called SecretSanta, operating with a simple logic that allows the users to create diverse pipelines and pipeline modifications.”
|
The origins of SecretSanta “I came up with this two-part name for the package, where (Secret) comes from the fact that the package is dealing with secreted proteins; the second part (Santa) was attached to indicate the fact that the package is very thoughtful and nice to the end user and presents clean and clear struggle free results.” Dr Anna Gogleva, SLCU Research Associate |
Dr Gogleva and her SLCU colleague Dr Hajk-Georg Drost, SLCU Research Associate and member of the Paszkowski research group who helped to further develop, scale and test SecretSanta, are witnessing an exponential rise in the amount of data being generated through the rapid sequencing of plant and animal genomes – all of which need to be processed and analysed. Dr Drost explains: “To keep up with analysing the huge amount of data that is now being generated through low-cost genome sequencing, we need something that brings together disparate tools into one comprehensive system. The aim of SecretSanta is to simplify these processes. SecretSanta implements tailored pipelines that can be applied to large protein sets to facilitate comparison of secretomes across multiple species or under various conditions.”
Technical specifications
The SecretSanta package provides an R interface for the integrative prediction of extracellular proteins that are secreted via classical pathways. It provides wrapper and parser functions around existing command line tools for prediction of signal peptides and protein subcellular localisation. The functions are designed to work together by producing standardised output. This allows the user to pipe results between individual predictors easily to create flexible custom pipelines and also to compare predictions between similar methods. To speed-up processing of large input fasta files, initial steps of the pipeline are automatically run as a massive parallel process when the number of input sequences exceeds a certain limit.
Read about SecretSanta in Bioinformatics.
SecretSanta is available as an open source package on GitHub.