Biological evolution
I am a theoretical biologist interested in the evolution of novelty. Since 2021, I am a research associate at the Sainsbury Laboratory, University of Cambridge (UK).
I like to think of life as organised into many interconnected levels: complex reaction networks make cells, many cells make organisms, organisms interact and form ecosystems. My main research question is: How did this multi-level complexity evolve?
To answer this question I develop mathematical models of evolution. My research focuses on three topics: the Origin of Life, microbial eco-evolutionary dynamics, and the transition to multicellularity.
Evolution of novelty
One of the most striking features of biological evolution is its endless ability to generate novelty. Novel traits, functions, shapes and forms have arisen throughout the entire 4 billion year history. I find the emerge of novelty fascinating - and deserving of an explanation. To do this, I make computational models of populations of cells that can interact with each other, replicate and mutate their genetic information. With these models I can simulate long evolutionary dynamics. Over long evolutionary time scales, cells evolve to integrate a large amount of information from their environment and can also evolve how to use it. That is, they evolve novel ways to behave and interact.
Evolution of gene regulation to integrate environmental information
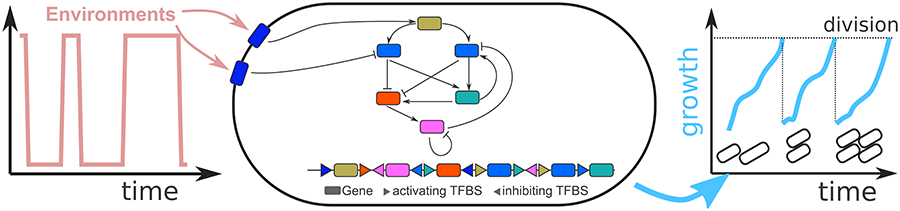
But how is this information integrated? And how does this depend on the type of environment cells are exposed to? For instance, if the environment fluctuates unpredictably - do we expect cells to adapt and re-adapt to each new environment? Or does a precise form of gene regulation evolves so that a cell can quickly steer its behaviour to match the environment? If the environment changes periodically, e.g. between day and night - can an internal clock evolve so that cells can anticipate the change?
To answer this question I am currently developing a model that allows cells to evolve their regulation in response to the environment.
Evolution of multicellularity – emergent multi-level evolutionary dynamics
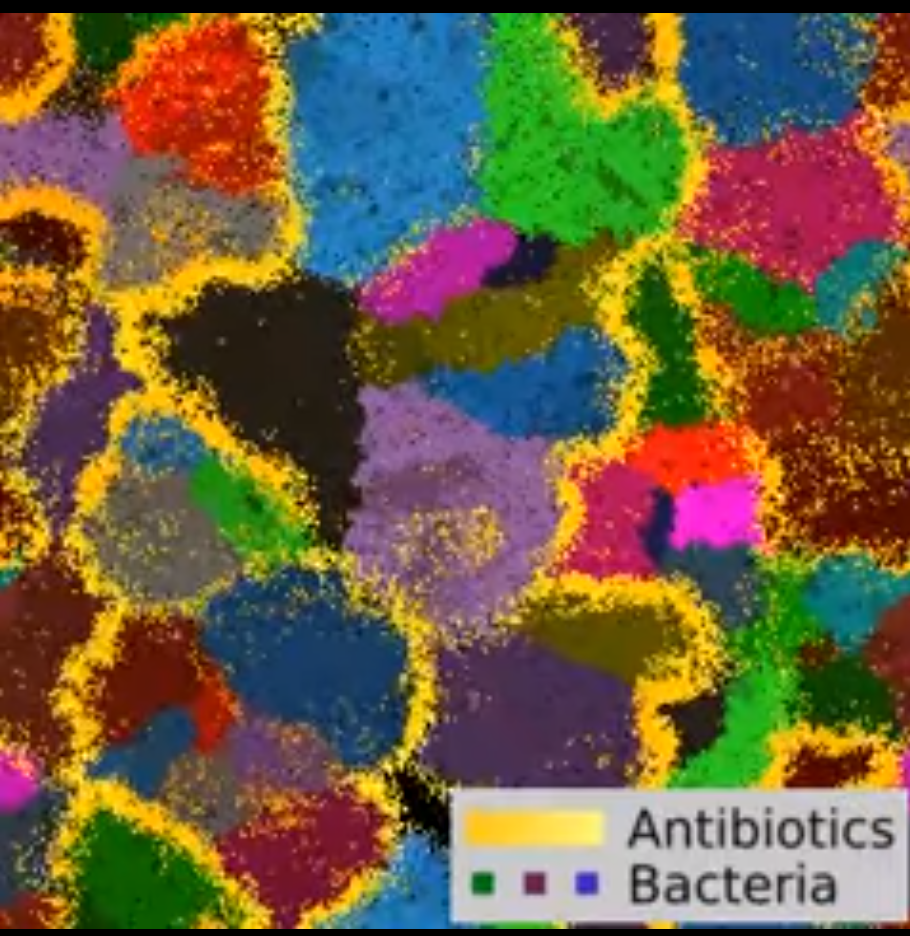
One of my long-term interest is the evolution of multicellularity. Many plants, animals and fungi and even some bacteria are multicellular, but their ancestors were not. How does a unicellular ancestor evolve into a differentiated multicellular organism? How does the infomation to generate different cell types get integrated in the genome? Following some recent experiments, I have made a model of the multicellular bacterium Streptomyces Coelicolor. The model shows that the competition between bacterial colony leads to a division of labour within the colony, in which some cells produce antibiotics and some others specialise in replication. The surprising insight of the model is that in order to divide labour, these bacteria must organise their genome in a precise way, so that mutations occur at the right genomic locations to allow cell differentiation.
Computational model of streptomyes colonies (colored according to descent) replicate and produce antibiotics (in yellow). Between-colony competition ensues when antibiotic halos touch. Watch the video
How does evolution generate the coordinated differentiation strategies that we observe today in multicellular development? And how does it differ from the phenotypic strategies of unicellular ancestral ecosystems? We made a computational model of cells that had to find resources to survive in a periodically changing environment. We showed that as multicellularity evolves, the strategy for finding these resources changes, because cells that previously lived as unicellular organisms, evolve to take each other into account in the multicellular cluster. As a bonus, we found that the patterns of differentiation of the multicellular cluster looked surprisingly similar to developmental dynamics of animals.
Multicellular clusters migrate along a chemoattracting gradient, and cells within the cluster replicate - generating protodevelopmental patterns. Watch video