Plant Developmental Evolution
Madelaine Bartlett
Research Group Leader
Sainsbury Laboratory
University of Cambridge
Bateman Street
Cambridge CB2 1LR
Email: mb2705@cam.ac.uk
Join the Bartlett Lab
If you're a prospective graduate student or a postdoc or wanting to work in the Bartlett Lab, please get in touch with Madelaine Bartlett via email at mb2705@cam.ac.uk.
Plant Evo-Devo
We are interested in the molecular underpinnings of plant diversity. Which genes control the development of plants? And how have these genes changed over the course of evolution?
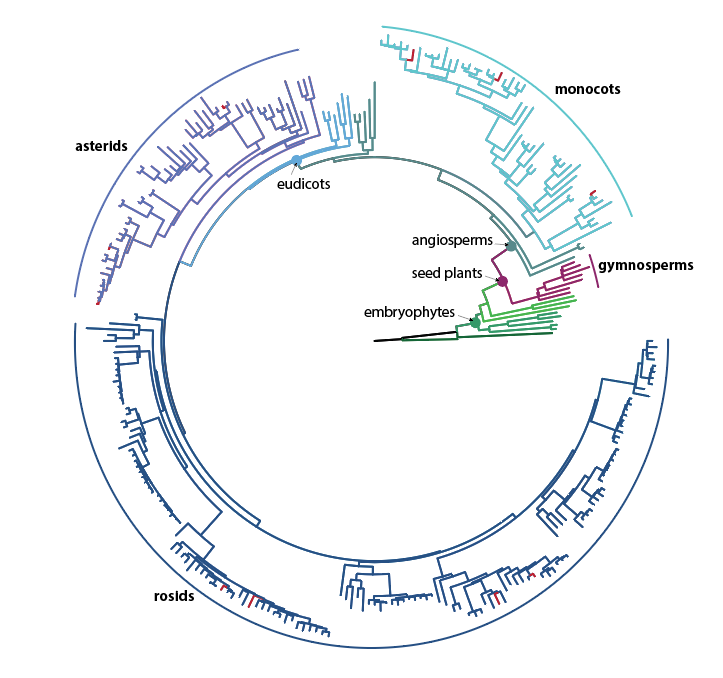
We focus specifically on the grasses to answer these fundamental questions about the genetic basis of plant development and morphological evolution. This remarkable family has diversified to occupy almost every terrestrial niche, and dominates many ecosystems. Multiple grass experimental systems allow us to dissect the molecular underpinnings of this diversity, and to reconstruct molecular and morphological evolution of key genes and traits.
Grass - the world's most important crop
There are about 11,000 species in the grass family Poaceae, making it the 5th largest family of plants. It is also one of the most diverse, with a huge variety in morphology, physiology, genetics and ecological habitats across the family. Nearly 70% of agricultural land is planted with crops from the grass family. Grasses include all the major cereals such as wheat, maize, barley and oats; pastures for livestock; sugar cane and woody species like bamboo. There is increasing interest in the minor grains like rye and the small seeded grasses known as millet, as well as tracing back the ancestors of our modern cereals to find forgotten genes that could benefit agriculture under a more highly variable climate.
Focusing on grasses means that our work has translational outcomes in crop improvement.
Mechanisms of morphological diversification in the grasses
We use quantitative phenotyping, genetics, genomics, and comparative phylogenetic methods to dissect the genetics of development, and to discover how molecular evolution impacts the evolution of plant form. Despite immense diversity, plants are constructed from homologous organs, using deeply conserved developmental genes. Understanding how the processes of evolution remixed, refashioned, and recast these conserved components to generate diversity will provide transformative blueprints and strategies for innovation in synthetic biology, crop engineering, and biomimetics.
To get there, we must first understand how complex morphological traits arise and change over deep time. We must also understand how evolutionary change to deeply conserved regulators affects development.
My research program is focused on identifying the genes that regulate the development of complex traits, defining how these genes function together in development, and determining how evolutionary change in these genes affects the evolution of organismal form. We integrate genetics, genomics, molecular evolution, and machine learning-enabled quantitative phenotyping to provide an integrative and multi-scale view of evolution. The grass traits we focus on are the evolution and development of floral sexuality, and the evolution and development of organ elaborations called awns. Both floral sexuality and awns directly impact plant fitness and yield. Thus, our work both reveals the fundamentals of organismal evolution, and has important implications in agriculture.
The evolution of conserved non-coding sequences in green plants
In a collaborative project with Idan Efroni (Israel), David Jackson (CSHL), and Zachary Lippman (CSHL, HHMI), we are working to identify deeply conserved cis-regulatory elements (CREs), including those that regulate developmental gene function, and to dissect the function of these CREs. As part of this work, we developed the ‘Conservatory’ algorithm, which identified cis-regulatory elements retained over deep time, detectable as conserved non-coding sequences (CNSs). The Conservatory database is available for you to explore.
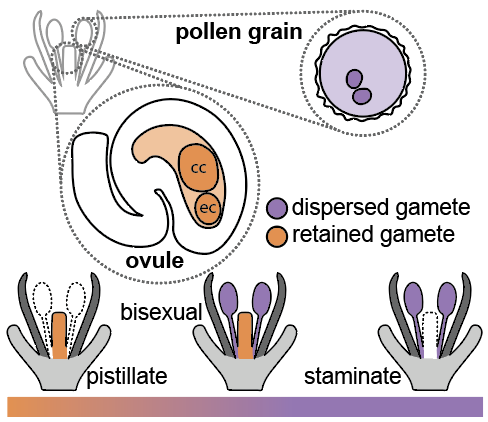
The development and evolution of floral sexuality
Most flowering plants do not have genetic sex determination. Instead, they have developmental sex determination, where floral organ development helps determine gamete identity. Almost all flowers are built of the same four floral organ types: sepals (called paleas in the grasses), petals (called lodicules in the grasses), stamens, and pistils. Pistils, which develop into fruits after fertilization, produce ovules which contain two gametes - the egg and central cells. Stamens produce pollen grains which contain two sperm cells. Thus, the type of gametes formed – egg and central cell, or sperm cells – is determined by patterns of pistil and stamen development. Most flowers produce both pistils and stamens (‘bisexual’), but strictly pistillate and staminate flowers have arisen many times over the course of evolution, with consequences to gene flow, reproductive success, and crop yield. In collaboration with Sam Leiboff (OSU), we work to determine the mechanisms underlying the development of unisexual flowers, and the evolution of diversity in floral sexuality, with a specific focus on the grass family.
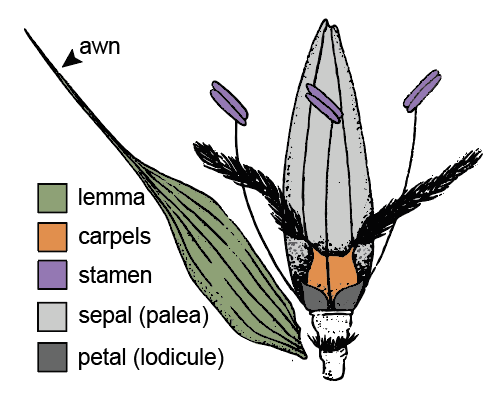
The development and evolution of awns
Many grasses have specialized organ elaborations called awns that can function in grain development, grain dispersal, and seedling establishment. Awns usually extend from leaf homologs called lemmas. Although often simple and needle-like, awns can take on elaborate forms, and can have many roles. For example, many twisted and bent awns are hygroscopic. These awns twist and untwist as water content changes, burying seeds deeper underground, which can help with seedling establishment. Other awns are long and feathery, and may aid in wind dispersal. Yet others, like those of barley and wheat, contribute photosynthate to developing seeds and impact grain weight. Thus, awns are diverse in form and function, and can contribute to grass success. In collaboration with Annis Richardson (University of Edinburgh) and Dana MacGregor (Rothamsted Research), we work to dissect the genetic underpinnings of awn development and evolution.