Alexander Jones
Research Group Leader
Sainsbury Laboratory
University of Cambridge
Bateman Street
Cambridge CB2 1LR
Plant hormones in action
The Jones team at the Sainsbury Laboratory Cambridge University (SLCU) is fascinated by how plants respond to the world. Stems and leaves grow towards light, whilst their roots grow towards water and nutrients. Some plants reproduce when triggered by stimuli such as warmth and day length. They reprogram their physiology when water is scarce and activate antimicrobial immune systems when they are being attacked by pathogens. Yet plants do not have a nervous system, a brain, or any of the sensory organs that we are familiar with in animals – so how do they know how to respond?
Cellular hormone dynamics
A big part of the answer lies in plant hormones. The term ‘hormone’ can simply refer to a chemical that acts as a ‘messenger’ within an organism, often travelling from one place to another to tell a particular cell what to do. In animals, these are often produced in special glands and target specific cell types, but in plants they are produced by a much wider range of cells and can target many cell types. With such distributed ‘decision making’, how do specific and appropriate responses to these mobile small molecules arise? Our view is that internal signals and external cues encode information in cellular hormone dynamics, and then cellular hormone signalling processes this information to activate appropriate downstream responses.
For instance, production of the plant hormone gibberellin (GA) can be triggered by cues like decreased light, increased temperature or salt stress. GA levels also fluctuate depending on internal cues, such as the stage at which the plant is in its lifecycle. Once GA accumulates to a certain level, it can trigger growth programmes. The exact nature of the growth that follows depends on the GA concentration, the plant tissue, and the timing.
The Jones group aims to map spatiotemporal hormone patterns at high-resolution in vivo and then reveal how specific hormone dynamics connect upstream cues and signals with downstream programmes relevant to development.
Biosensor engineering
In order to visualize these crucial cellular hormone dynamics, the Jones team has developed new high-resolution fluorescent biosensors based on initial developments when Dr. Jones was a postdoc in Professor Wolf Frommer’s lab. Gibberellin Perception Sensors (GPS1, GPS2, GPS3) are FRET based biosensors that are being used to unravel how cellular GA dynamics are determined as well as which aspects of these cellular GA dynamics are functionally relevant for GA dependent growth and development such as seed germination and root growth, apical hook development, and root symbioses. The Jones team has also recently developed additional FRET biosensors for other important plant hormones. Next-generation Abscisic Acid Concentration and Uptake Sensor 2 (ABACUS2) provides unprecedented resolution in our understanding of cellular ABA dynamics and allowed us to detect inter-organ growth coordination in response to humidity fluctuations. Additional sensors still in development include entirely novel FRET biosensors for auxin (IAA Genetically encoded Optical biosensors, IAAGO1 and IAAGO2) and the immune hormone salicylic acid (Salicylic acid Sensor, SalicS1).
The makings and meanings of cellular hormone dynamics
Our biosensors open new views into cellular hormone dynamics and launch many new questions. How are the patterns controlled and which aspects of the patterns are important? We found that GA levels in roots correlate with absolute cell elongation (Rizza et al. 2017 Nature Plants), however theoretical work by Leah Band and colleagues had suggested that GA would be correlated with relative cell elongation. We worked iteratively between our high-resolution experiments and Leah’s advanced multiscale mathematical models to identify key steps controlling cellular GA levels and new views on the quantitative relationship between GAs and growth (Rizza et al. 2021 PNAS) that we are currently investigating. We also leverage our high-resolution insights to collaborate with Henrik Jönsson (SLCU) to generate morphodynamic models that explain how hormones and other signals integrate with biophysical parameters to direct differential growth in stems. In our biosensor research it became clear that high-resolution sensing is not sufficient - we also need to be able to perturb hormones in high-resolution to distinguish correlation from causation. Thus, we engineered another enabling technology - an optogenetic gene expression switch for plants (‘Highlighter’).
Our future research aims to deploy biosensors, optogenetics and mathematical modeling to be able to mechanistically connect specific environmental conditions with their responses in plant physiology and development. This highly resolved view of information processing via hormones can then be applied to improve outcomes in horticultural and agricultural settings where manipulation of plant hormones, despite undesirable side-effects, has long contributed to global food security.
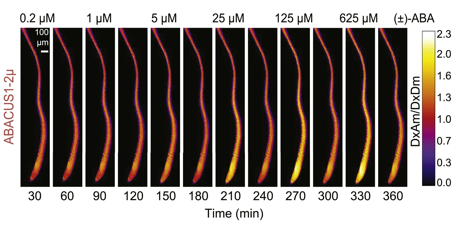
ABA responses of ABACUS1 in Arabidopsis roots: ABA responses of ABACUS1 in Arabidopsis roots. Images show fluorescence ratio (DxAm/DxDm) images showing pattern of ABACUS1-2µ in response to six pulses of increasing concentration of (±)-ABA. Right: look up table used for false coloring of ratio images. http://dx.doi.org/10.7554/eLife.01741.010
Key Publications
Tang B., Lu, J., Leontovyčová, H., Hoffmann, G., Rowe, J.H., Fouquay O’Donnell, S, Grangé-Guermente, M., Larsen, B., Wimalasekera, R., Carella, P., Incarbone, M., Kalachova, T., Jones, A.M. (2025) SALICYLIC ACID SENSOR1 reveals the propagation of an SA hormone surge during pathogen advance. Science accepted.
Kiradjiev, K.B., Griffiths, J., Jones, A.M.*, Band, L.R.* (2025) A balance of metabolism and diffusion articulates a gibberellin hormone gradient in the Arabidopsis root. PNAS revised. DOI: 10.1101/2025.03.24.643304
Dao, T. Q., Drapek, C., Jones, A.M., Leiboff, S. (2025) Comparing hormone dynamics in cereal crops via transient expression of hormone sensors. Plant and Cell Physiology. DOI: 10.1093/pcp/pcaf080
Kubalová, M., Griffiths, J., Müller, K., Jones, A.M., Fendrych, M. (2025) Gibberellin-deactivating GA2OX enzymes act as a hub for auxin-gibberellin crosstalk in Arabidopsis thaliana root growth regulation. PNAS. DOI: 10.1073/pnas.2425574122.
Rowe, J.H., Josse, M., Tang, B., Jones, A.M. (2025) Quantifying Plant Biology with Fluorescent Biosensors. Annual Review of Plant Biology. DOI: 10.1146/annurev-arplant-061824-090615
Zhang, Y., Anfang, M., Rowe, J.H., Rizza, A., Bar, H., Charrier, L., Geisler, M., Jones, A.M., Shani, E. (2025) ABA importers ABCG17 and ABCG18 redundantly regulate seed size in Arabidopsis. Plant Journal. DOI: 10.1111/tpj.70096
Van Praet, S., Rizza, R., Tang, B., Xie, C., Jones, A.M., Inzé, D., Vanholme, B., Depuydt, S. (2025) Coumarin Promotes Hypocotyl Elongation in Light-grown Arabidopsis thaliana Seedlings by Enhancing Brassinosteroid Signalling in an Auxin-Dependent Manner. Journal of Plant Growth Regulation. DOI: 10.1007/s00344-024-11617-z
Walia A.†, Carter R.†, Wightman R., Meyerowitz E.M., Jönsson H.*, Jones A.M.* (2024) Differential growth is an emergent property of mechanochemical feedback mechanisms in curved plant organs. Developmental Cell. DOI: 10.1016/j.devcel.2024.09.021
Griffiths J., Rizza A., Tang B., Feng L., Frommer W.B., Jones A.M. (2024) GIBBERELLIN PERCEPTION SENSOR 2 reveals genesis and role of cellular GA dynamics in light-regulated hypocotyl growth. Plant Cell. DOI: 10.1093/plcell/koae198
Drapek C., Rizza A., Mohd-Radzman N., Schiessl K., Dos Santos Barbosa F., Wen J., Oldroyd G.E.D.*, Jones A.M.* (2024) Gibberellin dynamics governing nodulation revealed using GIBBERELLIN PERCEPTION SENSOR 2 in Medicago truncatula lateral organs. Plant Cell. DOI: 10.1093/plcell/koae201
- Highlighted by: Jhu and Irving (2024) Seeing hormones in action: High-resolution gibberellin dynamics in nodules. The Plant Cell. DOI: 10.1093/plcell/koae216
- Highlighted by: Hurst and Gifford (2024) In preprints: hormonal stepping stones to diverging root organogenesis. Development. DOI: 10.1242/dev.202636
Lorrai, R., Erguvan, Ö., Raggi, S., Jonsson, K., Široká4, J, Tarkowská, D., Novák, O, Jones, A.M., Verger, S., Robert, S., Ferrari, S. (2024) Cell wall integrity modulates a PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) – HOOKLESS1 (HLS1) signalling module controlling apical hook formation in Arabidopsis. Plant Physiology. DOI: 10.1093/plphys/kiae370.
Rosa-Diaz I, Rowe JH, Cayuela-Lopez A, Arbona V, Diaz I, Jones AM (2024) Spider mite herbivory induces an abscisic acid-driven stomatal defense. Plant Physiology. DOI: 10.1093/plphys/kiae215
- Highlighted by: Gonzáles-Fuente (2024) Hold the door! Stomatal defense also protects against mites. Plant Physiology. DOI: 10.1093/plphys/kiae212
Larsen, B., Hofmann, R., Camacho, I.S., Clarke, R.W., Lagarias, J.C., Jones, A.R., Jones A.M. (2023) Highlighter is an optogenetic actuator for light-mediated, high resolution gene expression control in plants. PLOS Biology. DOI: 10.1371/journal.pbio.3002303
Rowe, J., Grangé-Guermente, M., Exposito-Rodriguez, M., Wimalasekera, R., Lenz, M., Shetty, K., Cutler, S.R., Jones, A.M. (2023) Next-generation ABACUS biosensors reveal cellular ABA dynamics driving root growth at low aerial humidity. Nature Plants. DOI: 10.1038/s41477-023-01447-4.
- Nature Plant Research Briefing: Visualising abscisic acid dynamics at low humidity in planta using ABACUS2. DOI: 0.1038/s41477-023-01469-y
Shi, B., Felipo-Benavent, A., Cerutti, G., Galvan-Ampudia, C., Jilli, L., Brunoud, G., Mutterer, J., Sakvarelidze-Achard, L., Davière, J-M., Navarro-Galiano, A., Walia, A., Lazary, S., Legrand, J., Weinstein, R., Jones, A., Prat, S., Achard, P.†, Vernoux, T.† (2024) A quantitative gibberellin signalling biosensor reveals a role for gibberellins in internode specification at the shoot apical meristem. Nature Comms. DOI: 10.1038/s41467-024-48116-4
Kubalová, M., Müller, K.,Dobrev, P.I., Rizza, A., Jones, A.M., Fendrych, M.† (2024) Auxin coreceptor IAA17/AXR3 controls cell elongation in Arabidopsis thaliana root by modulation of auxin and gibberellin perception. New Phytologist. DOI: 10.1111/nph.19557
Albuquerque-Martins, R., Szakonyi, D., Rowe, J., *Jones, A.M. and *Duque P. (2022), ABA signaling prevents phosphodegradation of the Arabidopsis SR45 splicing factor to negatively autoregulate inhibition of early seedling development. Plant Communications. DOI: 10.3390/life13061386
Mehra, P., Pandey, B.K., Melebari, D., Banda, J., Leftley, N., Couveur, V., Rowe, J., Anfang, M., De Gernier, H., Morris, E., Sturrock, C.J., Mooney, S.J., Swarup, R., Faulkner, C., Beeckman, T., Bhalerao, R.P., Shani, E., Jones, A.M., Dodd, I.C., Sharp, R.E., Sadanandom, A., Draye, X., Bennett, M. J. (2022) Hydraulic flux responsive hormone redistribution determines root branching. Science. 378(6621):762-768. DOI: 10.1126/science.add3771
Pablo Albertos, Tanja Wlk, Jayne Griffiths, Maria J Pimenta Lange, Simon J Unterholzner, Wilfried Rozhon, Theo Lange, Alexander M Jones, Brigitte Poppenberger (2022) Brassinosteroid-regulated bHLH transcription factor CESTA induces the gibberellin 2-oxidase GA2ox7, Plant Physiology, Volume 188, Issue 4, April 2022, Pages 2012–2025. DOI: 10.1093/plphys/kiac008
Robinson, S., Whitewoods, C., Otero, S., Jones, A.M. (2022) Vegetative Development Chapter. Plant Physiology and Development, 7th Edition.
James H Rowe, Annalisa Rizza, Alexander M Jones (2022) Quantifying Phytohormones in Vivo with FRET Biosensors and the FRETENATOR Analysis Toolset. In: Duque, P., Szakonyi, D. (eds) Environmental Responses in Plants. Methods in Molecular Biology, vol 2494. Humana, New York, NY. DOI: 10.1007/978-1-0716-2297-1_17
Martin Balcerowicz, Kartika N. Shetty, Alexander M. Jones (2021) Fluorescent biosensors illuminating plant hormone research, Plant Physiology, Volume 187, Issue 2, October 2021, Pages 590–602. DOI: 10.1093/plphys/kiab278
Cesar L Cuevas-Velazquez, Tamara Vellosillo, Karina Guadalupe, Hermann Broder Schmidt, Feng Yu, David Moses, Jennifer AN Brophy, Dante Cosio-Acosta, Alakananda Das, Lingxin Wang, Alexander M Jones, Alejandra A Covarrubias, Shahar Sukenik, José R Dinneny (2021) Intrinsically disordered protein biosensor tracks the physical-chemical effects of osmotic stress on cells. Nat Commun 12, 5438. DOI: 10.1038/s41467-021-25736-8
Rizza A, Tang B, Stanley CE, Grossmann G, Owen MR, Band LR, Jones AM (2021) Differential biosynthesis and cellular permeability explain longitudinal gibberellin gradients in growing roots. PNAS. DOI: 10.1073/pnas.1921960118
Jayne Griffiths, Roberto Hofmann, Alexander M Jones (2021) Signals| Gibberellin Signaling in Plants. Elsevier. DOI: 0.1016/B978-0-12-819460-7.00322-4
James H Rowe, Alexander M Jones (2021) Focus on biosensors: Looking through the lens of quantitative biology. Quantitative Plant Biology. DOI: 10.1017%2Fqpb.2021.10
Martin Balcerowicz, Kartika N Shetty, Alexander M Jones (2021) O auxin, where art thou? Nat Plants. DOI: 10.1038/s41477-021-00921-1
Annalisa Rizza, Alexander M Jones (2019) The makings of a gradient: spatiotemporal distribution of gibberellins in plant development. Current Opinion in Plant Biology. DOI: 10.1016/j.pbi.2018.08.001
Annalisa Rizza, Ankit Walia, Bijun Tang, Alexander M Jones (2019) Visualizing Cellular Gibberellin Levels Using the nlsGPS1 Förster Resonance Energy Transfer (FRET) Biosensor. J. Vis. Exp. (143). DOI: 10.3791/58739
Ankit Walia, Rainer Waadt, Alexander Jones (2018) Genetically encoded biosensors in plants: pathways to discovery. Annual Reviews. DOI: 10.1146/annurev-arplant-042817-040104
Rizza A, Walia A, Lanquar V, Frommer WB, Jones AM. In vivo gibberellin gradients visualized in rapidly elongating tissues. Nature Plants 2017. DOI: 10.1038/s41477-017-0021-9
- Recommended by F1000
- Highlighted in News & Views article: Azoulay-Shemer, T, Hsu P-K, Schroeder JI (2017) Nature Plants 3, 765–766. DOI: 10.1038/s41477-017-0025-5
Alexander M Jones (2015) A new look at stress: abscisic acid patterns and dynamics at high-resolution. New Phytologist. DOI: 10.1111/nph.13552
- Winner of the Tansley Medal. Lennon S. and Dolan L. (2016) New Phytologist. DOI: 10.1111/nph.13891
Alexander M Jones, Yuanhu Xuan, Meng Xu, Rui-Sheng Wang et al. (2014) Border control – a membrane-linked interactome of Arabidopsis. Science. DOI: 10.1126/science.1251358
Alexander M Jones*, Danielson JA, ManojKumar S, Lanquar V, Grossman G, Frommer WB* (2014) Abscisic acid dynamics in roots detected with genetically encoded FRET biosensors. eLife. DOI: 10.7554/elife.01741
- Highlighted in eLife Insight article: Choi W-G, Gilroy S. eLife. 2014; 3: e02763.
- Highlighted in The Scientist Modus Operandi article: Williams R. Stressing and FRETing. August 1st 2014.
Alexander M Jones†, Grossman G†, Frommer WB (2013) In vivo biochemistry: Applications for small molecule biosensors in plant biology. Current Opinion in Plant Biology. DOI: 10.1016%2Fj.pbi.2013.02.010 †equal contribution
Okumoto S†, Alexander M Jones†, Frommer WB. Quantitative imaging with fluorescent biosensors. Annu Rev Plant Biol. 2012. DOI: 10.1146/annurev-arplant-042110-103745 †equal contribution
Alexander Jones, Wildermuth MC (2011) The phytopathogen Pseudomonas syringae pv tomato DC3000 has three high-affinity iron-scavenging systems functional under iron limitation conditions but dispensable for pathogenesis. J Bacteriol. DOI: 10.1128/jb.00069-10
Gilkerson J., Hu J., Brown J., Jones A., Sun T.P., Callis J. (2009) Isolation and characterization of cul1-7, a recessive allele of CULLIN1 that disrupts SCF function at the C terminus of CUL1 in Arabidopsis thaliana. Genetics. DOI: 10.1534/genetics.108.097675
Alexander M Jones, Lindow SE, Wildermuth MC (2007) Salicylic acid, yersiniabactin, and pyoverdin production by the model phytopathogen Pseudomonas syringae pv tomato DC3000: synthesis, regulation, and impact on tomato and Arabidopsis host plants. J Bacteriol. DOI: 10.1128/jb.00827-07 *co-corresponding author, † equal contribution