Scientists at Sainsbury Laboratory have uncovered striking similarities in how two distantly related plants defend against pathogens despite splitting from their common ancestor more than 400 million years ago.
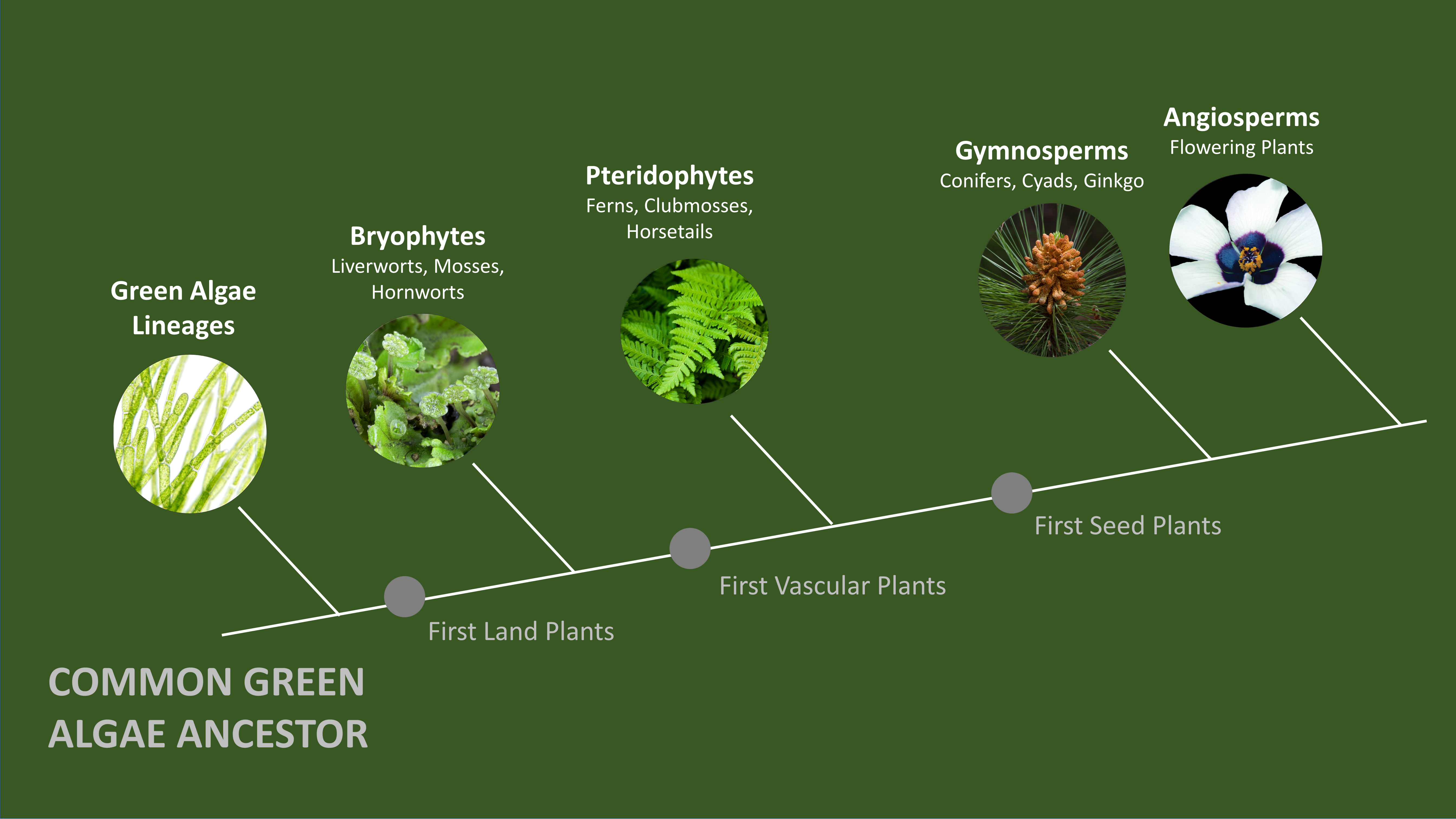
Our current understanding on how plants successfully defend against disease-causing pathogens mainly originates from studying economically important crop plants and a small number of closely-related flowering plant model systems. Very distantly-related plants, such as non-flowering liverworts that are believed to resemble some of the first land plants, are often overlooked. As a result, not much is known about how these plants defend themselves from pathogens.
 Researchers from the Schornack research team investigated how liverworts defend against an aggressive pathogen and compared their findings to a flowering plant – the first time such a comparison has been undertaken. By studying how these distantly related plants, which split from their common ancestor roughly 400 million years ago, respond to pathogen infections, the research team discovered microbe-responsive gene families that date back to early land plant evolution.
Researchers from the Schornack research team investigated how liverworts defend against an aggressive pathogen and compared their findings to a flowering plant – the first time such a comparison has been undertaken. By studying how these distantly related plants, which split from their common ancestor roughly 400 million years ago, respond to pathogen infections, the research team discovered microbe-responsive gene families that date back to early land plant evolution.
Published in Current Biology today, the identification of these evolutionarily conserved genes is shedding new light on the strategies that were likely critical for managing potentially harmful interactions with microbes during the expansion of plants onto land.
“We have shown that molecular responses to infection typical of modern flowering plants are common to very distantly-related land plants and may therefore be more ancient than we previously thought,” said Dr Sebastian Schornack, who led the research team that undertook the study. “Despite fluctuating environmental pressures over a broad evolutionary timescale, these conserved genes have retained their capacity to confer pathogen protection in plants, including in important agricultural crops.”
The team infected two distantly related plants – liverworts (Marchantia polymorpha) and wild tobacco (Nicotiana benthamiana) – with a filamentous pathogen (Phytophthora palmivora) to see what genes were upregulated (activated) in response.
“Infection with the pathogen triggered large changes in gene activities in both plants. We identified a subset of one-to-one corresponding genes (single-copy orthologs) in the liverwort and the wild tobacco and followed their level of activity during the infection.” said Anna Gogleva, co-author author and bioinformatics expert of this study. “Strikingly, some hallmarks of the infection response remained common between the two plants despite their large evolutionary distance.”
A number of different genes were activated in both plants, but a set of metabolic genes involved in phenylpropanoid (flavonoid) biosynthesis were highly activated in response to infection. These gene families are often associated with the stress-response in flowering plants, providing increased protection against biotic or abiotic stresses caused by chewing insects, pathogens and nutrient or light stress. However, this was the first time that these genes had been functionally linked to pathogen defence strategies in liverworts. These findings found further support thanks to a collaboration with Sara Stolze and Hirofumi Nakagami of the Max-Planck Institute for Plant Breeding Research (Cologne, Germany), who confirmed that proteins encoded by these genes are indeed differentially accumulating during infection.
To test whether these phenylpropanoid biosynthesis genes protect liverworts from pathogens the team needed plants where the genes involved in this biochemical pathway were inactive and overactive. They focused on a transcription factor protein that controls their gene activity, MpMyb14, which itself is induced during infection. Mutant liverworts which do not produce this transcription factor also did not activate the phenylpropanoid biosynthesis pathway in response to infection and were much more susceptible to disease. This indicated a critical role for phenylpropanoid metabolites in protecting liverworts from infection much like established plant-pathogen interaction principles in flowering plants.
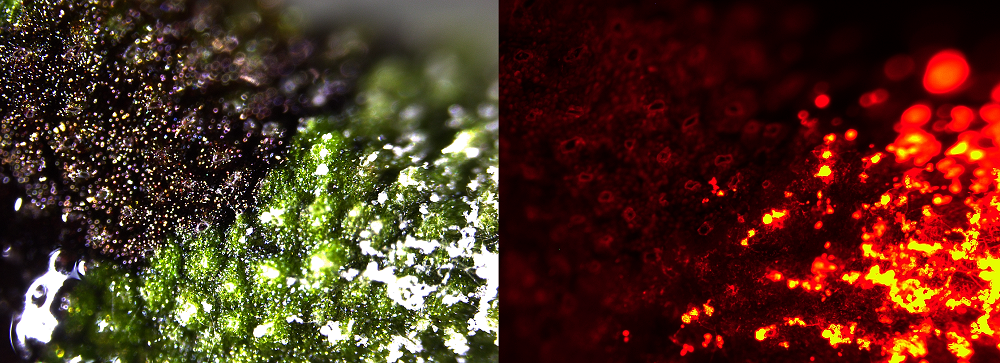
 “Pathogen zoospores germinate on the surface of liverworts and eventually colonise air chambers of the thallus and penetrate into photosynthetic cells, but in some areas we see an accumulation of a purple/red pigment and here the pathogen is rarely detected,” said Dr Philip Carella, lead author of the study. “We have evidence that demonstrates a key role for phenylpropanoid metabolism genes and the accumulation of pigment in providing liverworts with some resistance to pathogen infection.”
“Pathogen zoospores germinate on the surface of liverworts and eventually colonise air chambers of the thallus and penetrate into photosynthetic cells, but in some areas we see an accumulation of a purple/red pigment and here the pathogen is rarely detected,” said Dr Philip Carella, lead author of the study. “We have evidence that demonstrates a key role for phenylpropanoid metabolism genes and the accumulation of pigment in providing liverworts with some resistance to pathogen infection.”
The accumulation of the purple anthocyanin-like pigment is not all positive – plants with an overactive MpMyb14 at all times and across all tissues are stunted in their growth. To overcome this, the group produced plants that restrict the overactive MpMyb14 to some sections of the thallus and not others.
 “We produced liverwort plants with mosaic or sectorised pigment patterns – resembling military camouflage fatigues – that allowed us to compare resistance to pathogen infection in pigmented and non-pigmented areas of the same plant,” said Dr Carella. “It appears that the evolution of R2R3-MYB-mediated control of an expanding flavonoid/phenylpropanoid metabolite repertoire likely facilitated key advances in plant development (such as vertical growth) and stress physiology (antimicrobials, sunscreen molecules, antioxidants) that were essential for the expansion of plant life on land.”
“We produced liverwort plants with mosaic or sectorised pigment patterns – resembling military camouflage fatigues – that allowed us to compare resistance to pathogen infection in pigmented and non-pigmented areas of the same plant,” said Dr Carella. “It appears that the evolution of R2R3-MYB-mediated control of an expanding flavonoid/phenylpropanoid metabolite repertoire likely facilitated key advances in plant development (such as vertical growth) and stress physiology (antimicrobials, sunscreen molecules, antioxidants) that were essential for the expansion of plant life on land.”

The enormous diversity of traits and species that we see in modern plants today speaks to the millions of years of evolution that enabled plants to survive in dynamic and contrasting environments across the globe. “The conflict between organisms can be a very powerful selective pressure that guides their evolutionary trajectory,” said Dr Schornack. “Genes involved in fighting specific pathogens can evolve rapidly– both in plants and animals. But we also find these broadly-conserved genes responding to pathogen infection in very distantly-related plants, which suggests that land plants have retained a likely ancient pathogen deterrence strategy that is much too useful to lose”.
“Fossil evidence shows that plants have engaged in close-interactions with microbial life forms throughout their evolutionary history. Our research has now uncovered a common set of pathogen-responsive genes shared in early-divergent land plants and more evolutionarily young flowering plants. These genes have roles in cellular trafficking, transcriptional control, cell signalling, transport, and stress-associated metabolism and antimicrobial defences, which are all likely to have been critical for the expansion of plants onto land. Further comparative studies focusing on other distantly related land plants and their aquatic algal predecessors should reveal even more information about the evolution and role of these vital gene families.”
Journal article
Philip Carella, Anna Gogleva, David John Hoey, Anthony John Bridgen, Sara Christina Stolze, Hirofumi Nakagami, and Sebastian Schornack. (2019). Conserved Biochemical Defenses Underpin Host Responses to Oomycete Infection in an Early-Divergent Land Plant Lineage. Current Biology. DOI:https://doi.org/10.1016/j.cub.2019.05.078
Acknowledgements
This work was funded by the Gatsby Charitable Foundation, the Royal Society, the BBSRC OpenPlant initiative, the Natural Environment Research Council (NERC; NE/N00941X/1), and a Natural Sciences and Engineering Research Council of Canada (NSERC) postdoctoral fellowship to Philip Carella. Proteomic work performed in the Nakagami lab was supported by the Max-Planck-Gesellschaft.





