Cyanobacteria, an ancient lineage of bacteria that perform photosynthesis, have been found to regulate their genes using the same physics principle used in AM radio transmission.
New research published in Current Biology has found that cyanobacteria use variations in the amplitude (strength) of a pulse to convey information in single cells. The finding sheds light on how biological rhythms work together to regulate cellular processes.
In AM (amplitude modulation) radio, a wave with constant strength and frequency - called a carrier wave - is generated from the oscillation of an electric current. The audio signal, which contains the information (such as music or speech) to transmit, is superimposed onto the carrier wave. This is done by varying the amplitude of the carrier wave in accordance with the frequency of the audio signal.
The research team, led by Professor James Locke at the Sainsbury Laboratory Cambridge University (SLCU) and Dr Bruno Martins at the University of Warwick found that a similar AM radio-like mechanism is at work in cyanobacteria.
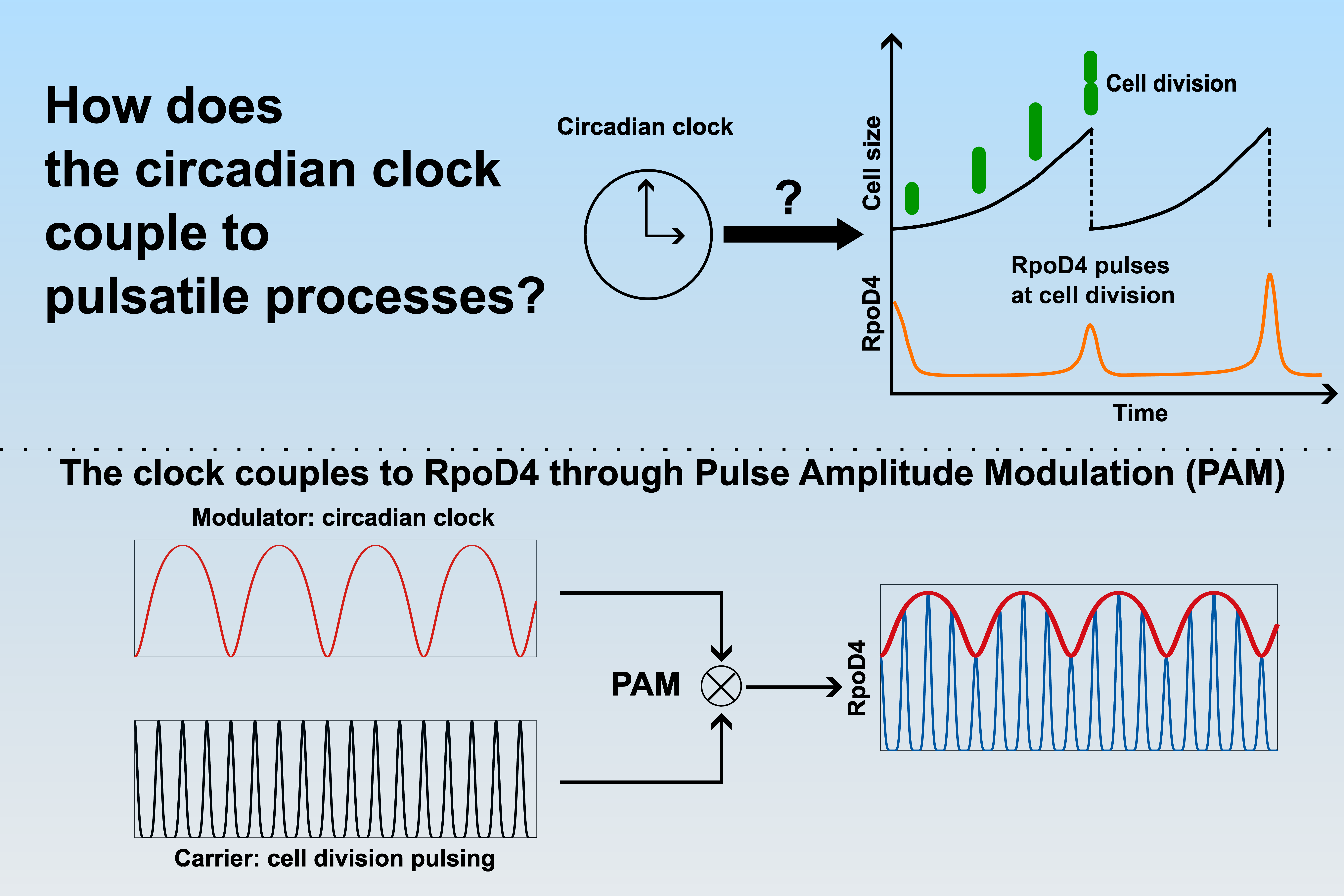
In cyanobacteria, the cell division cycle, the process through which one cell grows and divides into two new cells, acts as the ‘carrier signal’. The modulating signal then comes from the bacteria’s 24-hour circadian clock, which acts as an internal time-keeping mechanism.
This finding answers a long-standing question in cell biology – how do cells integrate signals from two oscillatory processes - the cell cycle and the circadian rhythm – which operate a different frequencies? Until now, it was unclear how these two cycles could be coordinated.
To solve the puzzle, the research team used single-cell time-lapse microscopy and mathematical modelling. With the time-lapse microscopy, they tracked expression of a protein, the alternative sigma factor RpoD4. RPoD4 plays an important role in the initiation of transcription, which is the process by which genetic information from DNA is transcribed into RNA. The modelling allowed researchersto explore signal processing mechanisms, comparing modeling results with microscopy data. The team found RpoD4 is turned on in pulses that occur only at cell division, which made it an ideal candidate for tracking.
Lead author Dr Chao Ye explained: “We found that the circadian clock dictates how strong these pulses are over time. Using this strategy, cells can encode information about two oscillatory signals in the same output: information about the cell cycle in the pulsing frequency, and about the 24-hour clock in the pulsing strength. This is the first time we've observed a circadian clock using pulse amplitude modulation, a concept typically associated with communication technology, to control biological functions.”
“Varying the frequency of either the cell cycle, through ambient light, or the circadian clock, through genetic mutations, validated the underlying principle. It is striking to see examples in nature of what we sometimes think of as ‘our’ engineering rules,” said co-corresponding author Dr Martins. “The cyanobacterial lineage evolved 2.7 billion years ago, and have an elegant solution to this information processing problem.”
Professor Locke added: “One reason we study cyanobacteria is that they have the simplest circadian clock of any organism, so understanding it lays the foundation we need to understand clocks in more complex organisms, like people and crops.
“These principles could have broader implications in synthetic biology and biotechnology. For example, this could help us develop crops that are more resilient to changing environmental conditions, with implications for agriculture and sustainability.”
Funding
This research was funded by BBSRC, ERC, the Gatsby Charitable Foundation and the Royal Society.
Reference
Chao Ye, Chris N. Micklem, Teresa Saez, Arijit K. Das, Bruno M. C. Martins, James C. W. Locke (2024) The cyanobacterial circadian clock couples to pulsatile processes using pulse amplitude modulation. Current Biology
DOI: https://doi.org/10.1016/j.cub.2024.10.047
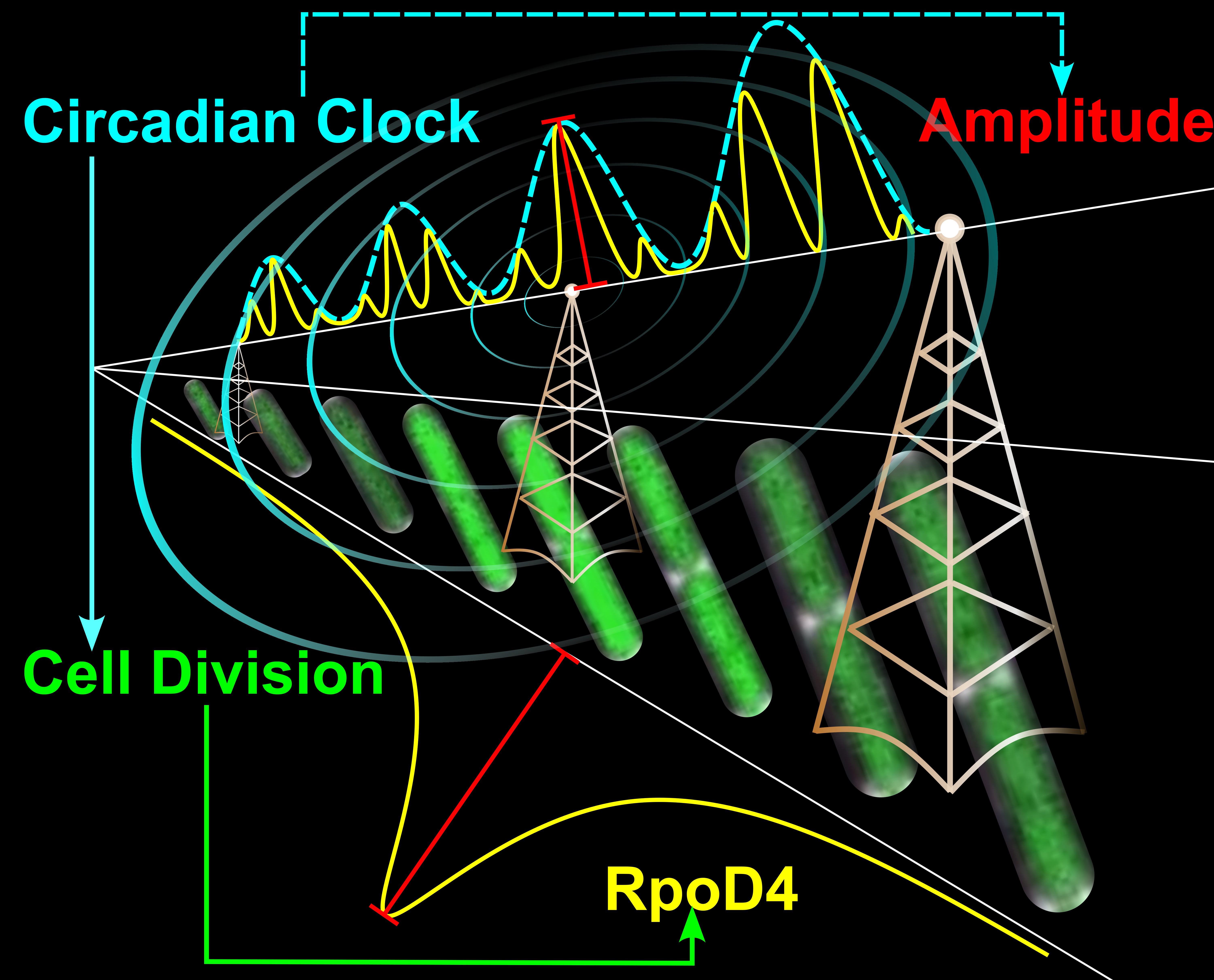
Graphic caption: At the centre, there are 3 radio towers (to indicate the analogy we used in the paper), and underneath the radio towers are WT cyanobacteria cells carrying the rpoD4 fluorescent reporter (from our time-lapse movie). As the cell grow and divide, the expression of rpoD4, as represented by the intensity of the reporter (in GREEN), increases and peaks at cell division. Below the rpoD4 reporter cells is a YELLOW trace quantifying the mean expression level of the rpoD4 reporter over time. The time trace peaks at cell division. The RED line with square ends indicates the amplitude of the rpoD4 expression pulse. Above the radio towers, there are time traces illustrating the Pulse Amplitude Modulation (PAM) mechanism. The dashed CYAN trace represents the modulating signal coming from the circadian clock, and the YELLOW traces illustrates rpoD4 pulses at different time points. No matter when the rpoD4 pulsing occurs, its amplitude is always under the modulation of the circadian clock. I also include a red line to show the amplitude of one rpoD4 pulse. At the centre, the CYAN radio waves emitting from the central radio tower is another illustration of the fact that the modulating signal comes from the circadian clock. At the edge of this graph, I listed all key components (circadian clock, cell division, RpoD4 and amplitude) of the PAM mechanism, and colour coded them based on their corresponding traces in the graph. I also use arrows to indicate the relationships between there components. Graphics by Chao Ye.