SLCU is delighted to welcome Dr François Nédélec to the join its research leadership team.
Dr Nédélec brings with him a wealth of knowledge and experience in cell and developmental biology with a multidisciplinary background in mathematics, physics and biology combining theoretical and experimental investigations to exploring cell biology dynamics.
He will establish a new research group at SLCU that will apply both physical and theoretical approaches to study the organisation of living cells, with an emphasis on the cytoskeleton.
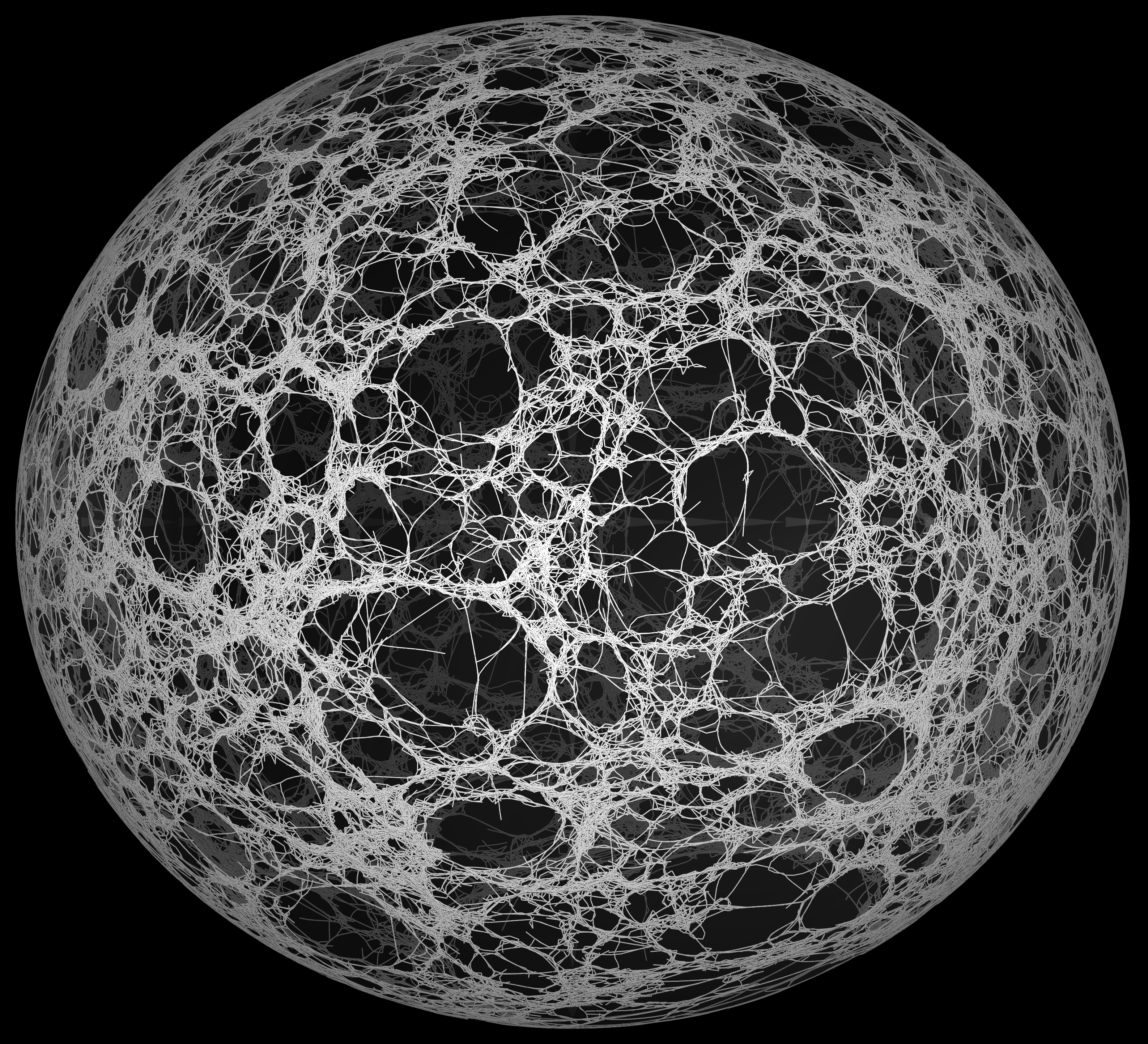
Dr Nédélec has led the development of a number of bio-physical and quantitative analysis methods, including establishing cytosim, an Open Source simulation project. The cytosim platform enables the study cytoskeletal systems from a mechanistic perspective, modelling the dynamics of networks involving microtubules or actin and associated proteins.
Dr Nédélec also has an interest in studying mitotic spindles, which are large cellular structures made for segregating the chromosomes during eukaryotic cell division. He previously studied mitotic spindle mechanics in a range of organisms (yeast cells and Xenopus) and will undertake further work using experimental and theoretical methods to analyse the organisation of spindles and other cytoskeletal structures in plants.
His long-term research objective is to understand physical principles associated with the organisation in living cells, and at a higher level for multiple cells interacting.
His next focus will be on looking at how plants overcome engineering problems when building their cells.
“I am interested in understanding the mechanical side of cell morphogenesis and how the cell shape confers advantageous mechanical properties,” Dr Nédélec said. “I want to look at how the cytoskeleton and cell wall interact as cells grow and divide, as well as exploring spindle and nuclear positions during cell development and replication.
“I am really excited to be joining SLCU, where I am establishing a team to continue research on the cytoskeleton, with an added focus on morphogenesis.
“Plant cells are surrounded by polymeric mesh called the cell wall, which dominates their mechanical properties. The cells themselves have turgor pressure and they push on the cell wall that constrains them, so there is always a lot of tension in plant tissues (this is why a fresh lettuce is crispy). But interestingly for us, cells use their cytoskeleton to direct the addition of new cell wall material. This is the perfect system to study how the cytoskeleton directs cell growth and cell division which together determine the shape of cells, and thus ultimately the shape of the whole plant.
“We are still a long way from being able to predict the shape of a plant organ, such as a flower, with this approach, but this is a stimulating aim that offers many valuable projects along the way.”
Dr Nédélec was most recently a visiting researcher at the Center for Interdisciplinary Research in Paris and prior to that he led a research group at EMBL between 2005-2017. He was also the co-founder of the EMBL mathematical modelling centre, which provided a training programme for biologists, seminars and help desk for researchers.
We asked Dr Nédélec about how he came to be a research scientist and his goals in science.
What got you interested in science in the first place?
I grew up spending much of my spare time in the basement of our home, building boats and airplanes out of wood. I also liked disassembling old computers and appliances to salvage the motors and other parts, and reassembling them into new things. If I was not in the basement, you might find me upstairs, in front of a first-generation Apple Computer Macintosh, where I wrote all sorts of little programs in Basic, Pascal, Forth and C. At this time, if I took apart some living thing, I was never able to put it back together, and I mostly stayed away from that.
My fascination with biology came much later.
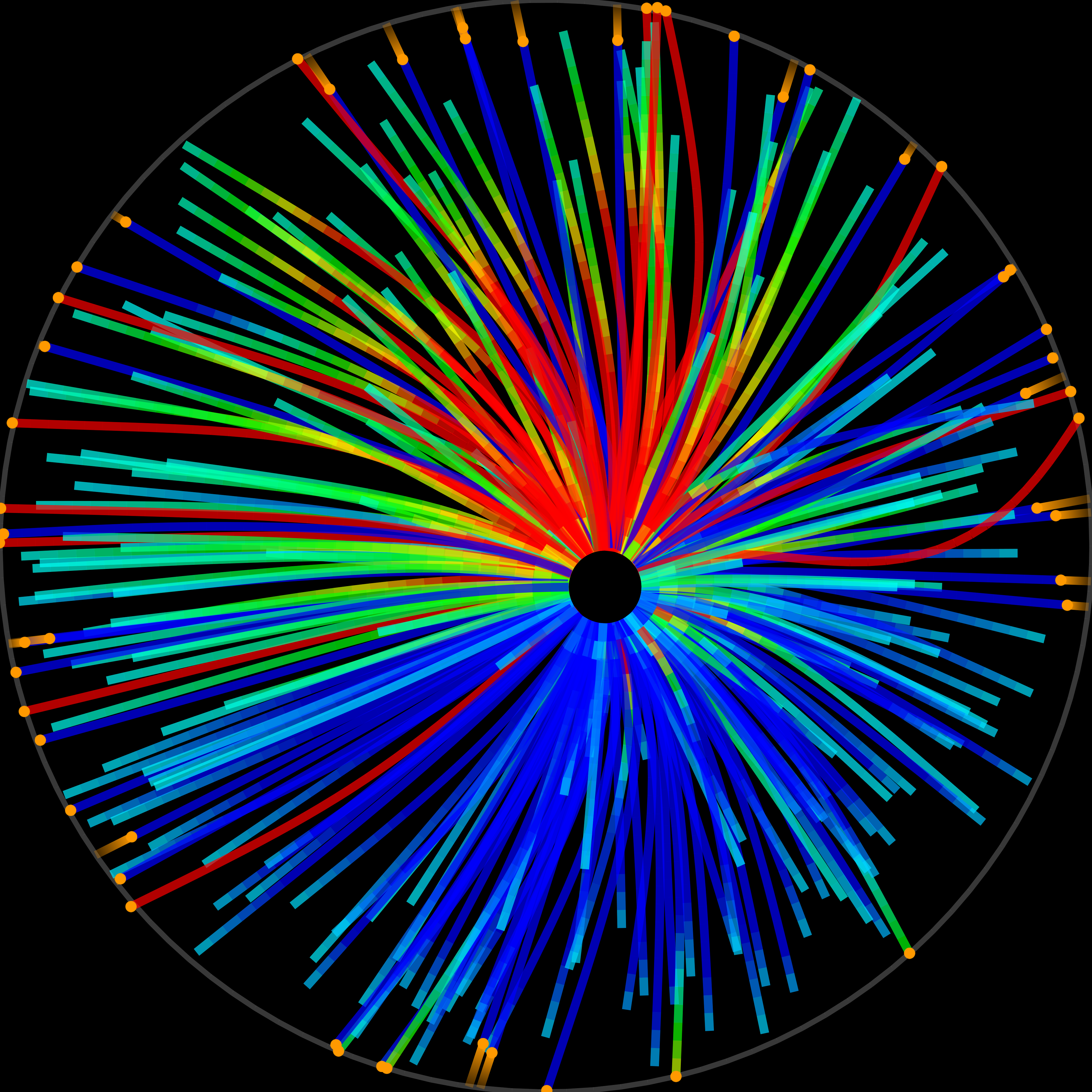
What was your first research topic?
It was an incredibly exciting time for the physical and biological sciences when I was looking to embark on my PhD – molecular motors had just been characterised! This was a major influence on me and I choose to do my PhD with Anthony C. Maggs on a topic at the interface between physics and biology.
I joined the lab of Stan Leibler at Princeton, where everyone was tinkering with the cytoskeleton. Mike Surette taught me some basic molecular biology, so I could purify kinesin, a molecular motor that could walk on microtubules... I remember spending a lot of time in the cold room but with music, a warm jacket and a clear goal in mind, which was not bad at all. With this kinesin I built a system that would self-organise into beautiful patterns.
It was the dawn of ‘Synthetic Biology’, but in the eyes of a physicist, I had created a dissipative system, which escaped the principles of equilibrium physics that we had learned in school. New theories would have to be developed to describe the phenomenon.
In the evenings, I went back to computers and wrote a program to reproduce the self-organization of the filaments and motors. At this time, it took a few days to simulate this system, but now it only takes a couple of minutes for the same job! At the end of my PhD, I was totally committed to studying Biology, where I could exert my life-long passions of tinkering and computer programming.
How did you develop your research after your PhD?
I became interested in understanding how filaments were organized and decided to see if I could study them in vivo, rather than in vitro, to maybe one day contribute to the understanding of living cells. This led me to joining the group of Eric Karsenti at the EMBL in Heidelberg.
I spent 20 years at EMBL where I built my first group. We studied diverse cytoskeletal processes, focusing on their mechanical and dynamical properties using a combination of experiments and simulations. We studied the formation of the mitotic spindle, the positioning of the spindle and also the movements of nuclei, which are processes that are typically associated with the microtubule cytoskeleton. The simulation became known as "Cytosim", and in 2012 it was completely restructured, becoming dramatically more modular and powerful.
Around 2014 we had developed Cytosim to enable it to study actin networks contraction, which is fundamental to understand morphogenesis in animal cells, and other actin-associated systems such as endocytosis or cytokinesis.
Why Cambridge?
I am really excited to be joining SLCU, where I am establishing a team to continue research on the cytoskeleton, with an added focus on morphogenesis.
Plant cells are surrounded by polymeric mesh called the cell wall, which dominates their mechanical properties. The cells themselves have turgor pressure and they push on the cell wall that constrains them, so there is always a lot of tension in plant tissues (this is why a fresh lettuce is crispy). But interestingly for us, cells use their cytoskeleton to direct the addition of new cell wall material. This is the perfect system to study how the cytoskeleton directs cell growth and cell division which together determine the shape of cells, and thus ultimately the shape of the whole plant.
We are still a long way from being able to predict the shape of a plant organ, such as a flower, with this approach, but this is a stimulating aim that offers many valuable projects along the way.





