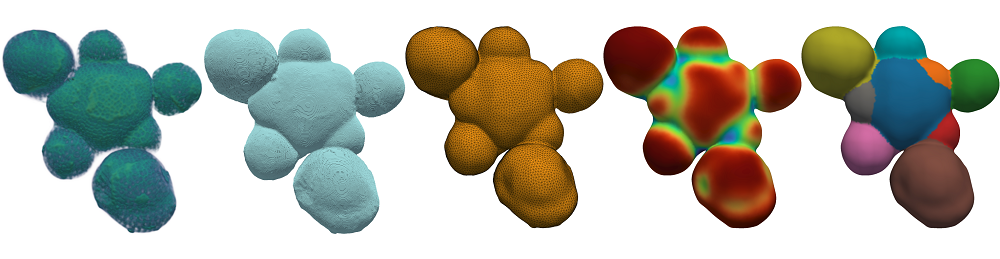
Illustration of the tissue-level segmentation workflow showing different processing stages, consisting of the 1) raw data, 2) contour, 3) surface mesh, 4) mesh curvature, and 5) the final, segmented mesh. From the segmented data, sub-tissue geometrical properties can be quantified.
New high-throughput method to quantify flower organ development
A new computational method that analyses plant tissues in 3D provides direct quantification of plant morphology at the tissue-level and promises to reveal phenotypes not easily identifiable by eye.
The growing shoot tips of plants, called meristems, are made-up of clusters of rapidly dividing stem cells that are the precursor cells from which all the different parts of a plant grow – stems, leaves, flowers and fruit. Plant scientists measure the observable traits (phenotype) of these critical tissues using confocal microscopes and a number of manual methods have been developed to quantify physical features in the meristem images.
Employing a computer to do the quantifications for you removes the risk of human error. Not only would you save time, the computer might even identify changes, for example new phenotypes resulting from genetic mutations, you would never have picked up by eye alone and, consequently, provide new insights into plant growth and development.
A method developed at the Sainsbury Laboratory Cambridge University (SLCU) accelerates and improves analysis of confocal imaging data to identify and quantify specific features (segmentation) in tissue-level structures such as shoot apical meristems and early flower organs. Key advantages of the method are that tissues can be kept intact as no preparatory chemical reagents are needed, it is applicable to low-resolution data, and a large number of samples can be analysed simply and quickly.
However, the method’s biggest benefit is that the semi-automatic workflow removes the risk of human-error caused by manual quantification techniques and qualitative assessment of tissue morphology by eye – making it a truly reproducible and unbiased method.
The method uses the surface curvature to identify and extract surface meshes on 3D plant tissues to identify patterns and organ-level features, which can then be used to determine phenotypes. The data also provides a potential new framework for classifying plant tissue development stages, and identifying the transition between them, such as in early flowers.
First author of the method published in Frontiers in Plant Science, Henrik Åhl, Research Associate in Professor Henrik Jönsson’s Group at the Sainsbury Laboratory Cambridge University (SLCU), said quantifying the shape of meristems provides insights into the regulatory systems behind shoot development and their stem cells. “We created a simple method for segmenting out early primordia based on the tissue curvature. This allows us to quantify geometric – and other – properties on an organ-by-organ basis using confocal microscope data,” he said.
“Shape and regulation go hand-in-hand, but we need quantitative data (and lots of it) to assess whether an apparent phenotype, such as a change in shape, really is a consequence of altered regulation. As plant scientists we normally rely on our eyes to determine phenotype changes, but this computational method will help us to identify phenotypes we wouldn't ever have been able to identify by eye.”
The geometric arrangement of organs, called phyllotaxis, is largely determined by how the hormone auxin is patterned in the shoot, however, this method has revealed new insights into phyllotaxis robustness.
“The positions of auxin maxima in the shoot specify where new organs are formed. There is a well-known, fundamental result: PIN1, an auxin transport protein, polarises in defined patterns in the shoot to guide auxin to these maxima. However, our results show how also auxin transport proteins not expressed in the shoot – especially PIN3/4/7 – affect the variability in organ positioning, showing how phyllotactic robustness is partly regulated on the plant level.
“We also show that our method can be used to identify regulators of shoot morphology that haven’t been described before. By looking at mutants with defective functionality of addition auxin transport proteins ABCB1/19, which are canonical long-range transporters of auxin, we see that both the shoot morphology and phyllotaxis are perturbed, especially for early organs. We also looked at GFP-tagged ABCB19, and found that it has a clear expression pattern in early primordia, implicating these auxin transporters as regulators of shoot morphology. Further work is needed, but it is possible that this helps amplify auxin flux from biosynthesis hotspots in early flowers.”
This new method has also provided new understandings into tissue-level morphology.
Åhl explains: “Plant cells are surrounded by stiff cell walls and the wall properties are important to determine the shape of a plant organ. By looking at a large set of wall perturbed plants for which expression domains are known, we found that cell wall biosynthesis genes which are expressed in the periphery of the shoot tend to prevent excessive flattening of the shoot. This suggests that differential expression domains for functionally redundant homologues could be important in tuning the tissue-level morphology.”
Åhl says this method is also a great way of identifying where the stem cell niche is located in plants that don’t have fluorescent markers, rendering a way of parameterising the plant shoot even with data of limited scope and quality.
This method will enable plant science researchers to address fundamental questions of plant development by quantitative phenotyping with high throughput, consistency and reproducibility.
Reference
Åhl, H., Zhang, Y. and Jönsson, H., High-throughput 3D phenotyping of plant shoot apical meristems from tissue-resolution data. Frontiers in Plant Science, p.712. https://doi.org/10.3389/fpls.2022.827147





