A team of Cambridge scientists working at the intersection between biology and computation has found that random gene activity helps patterns form during development of a model multicellular system.
We all start life as a single cell, which multiplies to produce specialised cells that carry out different functions. This complex process relies on precise controls along the way, but these new findings suggest that random processes can also play an important role during the development of living systems.
In research published today in Nature Communications, the scientists from James Locke’s team at Sainsbury Laboratory Cambridge University and collaborators from Andrew Phillip's team at Microsoft Research describe their surprising discovery of order in randomness while studying bacterial biofilms.
A biofilm develops when free-living single-celled bacteria attach to a surface and aggregate to start multiplying and spreading across the surface. These multiplying individual cells mature to form a three-dimensional structure that acts like a multicellular organism.
And while individual cells can survive on their own, these bacteria prefer to work together with biofilms being the dominant form found in nature. This biofilm consortium provides bacteria with important survival advantages such as increased resistance to environmental stresses, including antibiotics.
The researchers developed a new time-lapse microscopy technique to track how genetically identical single cells behave as the living biofilm developed. Dr Eugene Nadezhdin, joint lead-author, said: “We looked at how cells decide to take on particular roles in the biofilm. We found that towards the surface of the biofilm there were two different cell types frequently present – cells that form dormant spores and those that keep growing and activate protective stress responses. Although these two cell types are mutually exclusive, they both could exist in the same location.”
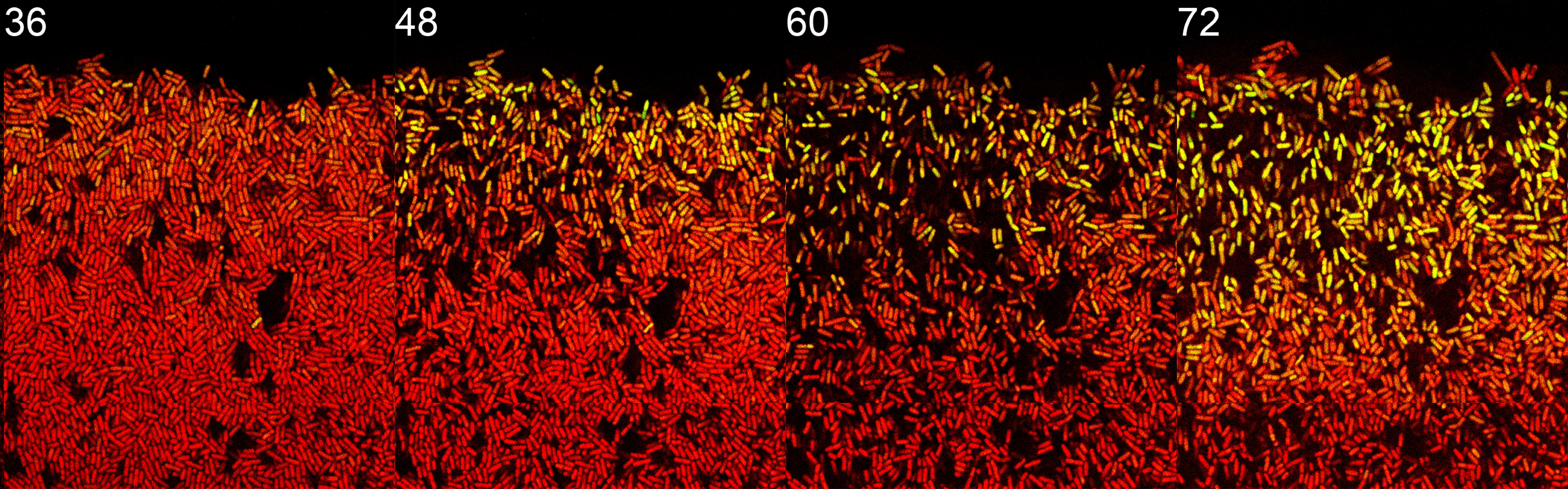
They focussed on obtaining a detailed picture of how gene expression (whether genes are active or inactive) changes over time for the individual cell types, specifically on expression of a regulatory factor, called sigmaB, which promotes stress responses and inhibits spore formation. They found that sigmaB randomly pulses on and off in cells at hourly intervals, generating a visible pattern of sporulating and stress-protected cells across the biofilm.
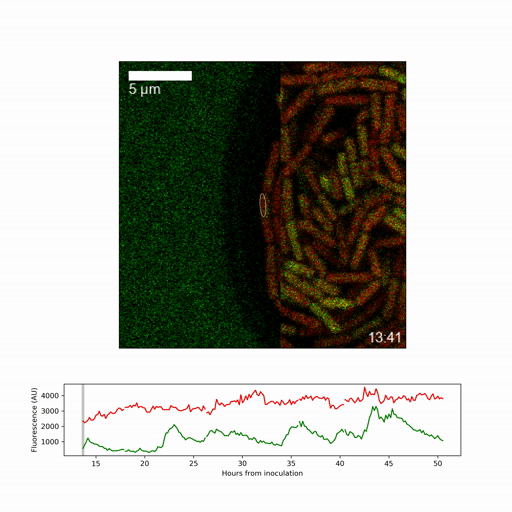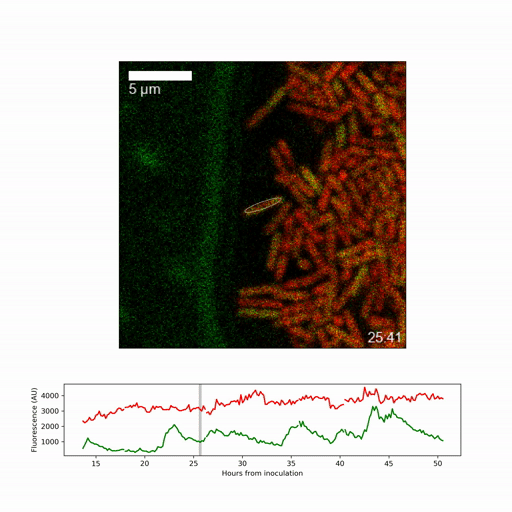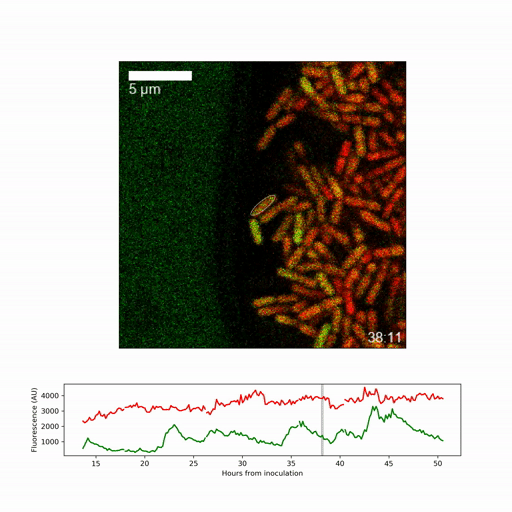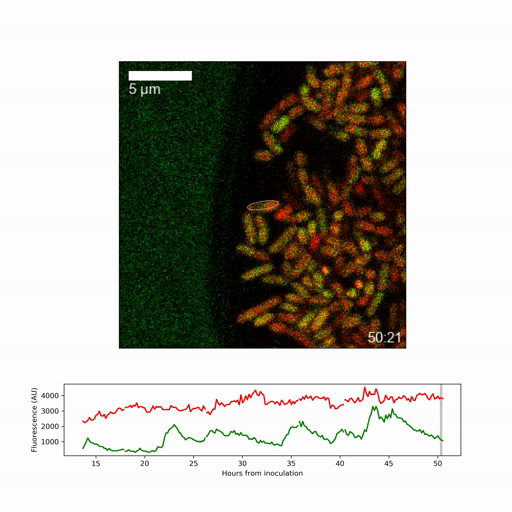
To understand the implications of the pulsing, the researchers generated a mathematical model of the sigmaB-controlled stress response and sporulation systems. Dr Niall Murphy, joint lead-author, said: “The modelling revealed that random pulsing enables, at any one time, only a fraction of cells to have high sigmaB activity and activation of the stress pathway, allowing the remainder of cells to choose to develop spores. While the pulsing is random, we were able to show through a simple mathematical model that increasing expression of the gene creates shifting patterns among the different regions of the biofilm.”
The results demonstrate how random pulsing of gene expression can play a key role in establishing spatial structures during biofilm development.
Dr Locke said: “This randomness appears to control the distribution of cell states within a population – in this case a biofilm. The insights gained from this work could be used to help engineer synthetic gene circuits for generating patterns in multi-cellular systems. Rather than the circuits needing a mechanism to control the fate of every cell individually, noise could be used to randomly distribute alternative tasks between neighbouring cells.”
According to Dr Phillips: "This use of noise as an allocation mechanism could have important implications for understanding how living cells share scarce resources, and could potentially be harnessed to develop more efficient genetic systems in the future, for a range of applications in biomanufacturing and therapeutics".
Reference
Eugene Nadezhdin, Niall Murphy, Neil Dalchau, Andrew Phillips and James Locke (2020) Stochastic pulsing of gene expression enables the generation of spatial patterns in Bacillus subtilis biofilms. Nature Communications. DOI: 10.1038/s41467-020-14431-9.






