Biography
Growing up on a farm near the Danube Valley in Austria, I had the freedom to explore wildflower meadows, orchards and forests from an early age. This childhood experience built the foundation for my life-long fascination for plants and the diversity we find among their organs in size, shape and function. Already back then, I was particularly fascinated by the odd-looking lumps and bumps we find growing on roots and leaves that we commonly refer as plant galls.
I studied Plant Sciences at the University of Applied Life Sciences of Vienna (BOKU, Wien) where I did my diploma project on resistance in wheat to Fusarium head blight, a fungus that can infect wheat plants specifically through floral organs at flowering time. During my PhD I worked on the regulatory and cellular mechanisms that underlie floral organ development under the supervision of Prof. Robert Sablowski at the Cell Developmental Biology Department at the John Innes Centre in Norwich, UK.
To expand on the question of how plant organs of specific sizes, forms and functions are generated the symbiotic root nodule in legumes caught my attention. I joined the group of Prof. Giles Oldroyd and the Enabling Nutrient Symbioses in Agriculture (ENSA) consortium as a Post-Doc. In my research I have focused on the overarching question of how the symbiotic root nodule has evolved in legumes and what gene regulatory networks underlie nodule organ identity using the model legume Medicago truncatula.
Research Interests
That fact that the development of specialised symbiotic root organs is induced in legumes in response to the colonising rhizobial bacteria and that these purpose-built plant organs enable intracellular accommodation of thousands and thousands of nitrogen-fixing bacteria fascinates me every single day.
Through my work, I have become more and more intrigued by the fact that parasitic and symbiotic plant colonisers as heterogenous as bacteria, fungi, nematodes and insects converge on similar organogenesis pathways to direct a diversity of purpose-built plant galls. Hence these colonisers can be considered as generators of plant organ diversity. Studying these interactions at molecular level helps us to advance and refine our understanding of the common principles that underpin plant organ diversification.
Using a combination of developmental genetics, imaging and genome-scale profiling approaches we are discovering and characterising the gene regulatory pathways that are recruited and modified by these colonising organisms to generate the unique features of plant galls such as special feeding sites and intracellular accommodation. I am particularly interested in obtaining quantitative data that can build the foundation for computational models explaining the morphogenesis and evolution of these novel and unique organs.
Furthermore, I am interested in the molecular means by which these colonisers access and modify these plant developmental pathways. For this question at the intersection between molecular plant-microbe interactions and plant development, I consider it important to foster collaborations with groups working on plant-pathogen interactions and effector biology.
Currently, we predominantly use the legume Medicago truncatula (barrel medic), a model for temperate legume crops such as pea, because it is a common host for nodule-inducing nitrogen-fixing bacteria and root-gall-inducing parasitic root-knot nematode species. This provides us with a framework to compare the plant developmental regulatory pathways of endogenous lateral roots, symbiotic root nodules and parasitic root galls within the same genetic background and tissue context.
Relating back to my agricultural background, I am very interested in the translatability of my research. Therefore, I anticipate that the outcomes of our research will not only advance our understanding of the common principles of plant organ diversification, but also have the potential to provide future strategies to improve the resilience of our crops against parasitic colonisers and to promote biological nitrogen fixation.
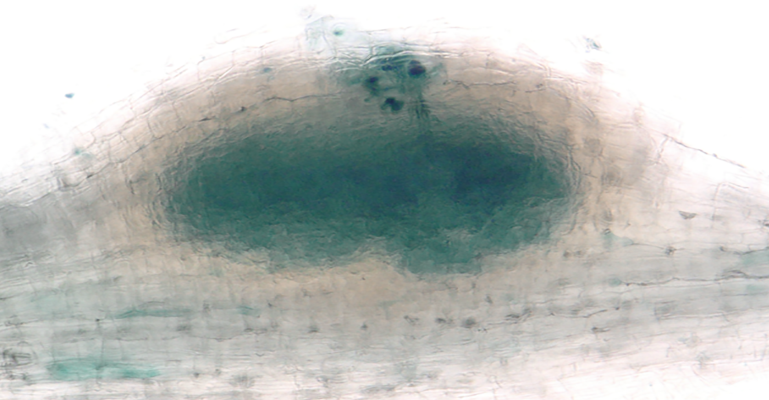
Medicago root nodule emerging from the primary root fully colonised with nitrogen-fixing beneficial bacteria 7 days after inoculation. Bacteria labelled in blue.
Research published in the media and science features
- Overlap in lateral root and nodule development brings self-fertilising cereals one step closer | Sainsbury Laboratory
- Growing excitement about self‑fertilising crops | The Times
- How petals shape up | PLOS Biologue
- How do cell measure themselves? | Tech Explorist
Publications
Tak Lee, Martina Orvosova, Morgane Batzenschlager, Marcelo Bueno Batista, Paul C. Bailey, Nadia A. Mohd-Radzman, Aram Gurzadyan, Naomi Stuer, Kirankumar S. Mysore, Jiangqi Wen, Thomas Ott, Giles E.D. Oldroyd, Katharina Schiessl (2024) Light-sensitive short hypocotyl genes confer symbiotic nodule identity in the legume Medicago truncatula. Current Biology. https://doi.org/10.1016/j.cub.2024.01.018
Colleen Drapek, Nadiatul Radzman-Mohd, Annalisa Rizza, Katharina Schiessl, Fabio Dos Santos Barbosa, Jiangqi Wen, Giles E.D. Oldroyd, Alexander M. Jones: Cellular gibberellin dynamics govern indeterminate nodule development, morphology and function. bioRxiv 2023 (https://doi.org/10.1101/2023.09.09.556959) PRE.
Schiessl K†, Lee T, Orvosova M, Bueno-Batista M, Stuer N, Bailey P C, Mysore K S, Wen J and Oldroyd G E D: Light sensitive short hypocotyl (LSH) confer symbiotic nodule identity in the legume Medicago truncatula. bioRxiv 2023 (https://www.biorxiv.org/content/10.1101/2023.02.12.528179v1) PRE.
Roy S, Jerez T, Zhang S, Liu W, Schiessl K, Boschiero C, Lee H-K, Zhao PX, Murray JD, Oldroyd GED, Scheible WR, Udvardi M: The peptide GOLVEN10 controls nodule and lateral root organogenesis and positioning along the longitudinal root axis. bioRxiv 2022 (https://www.biorxiv.org/content/10.1101/2022.05.06.490929v1) PRE.
Feng, J, Lee T, Schiessl K, and Oldroyd G E D: Processing of NODULE INCEPTION controls transition to nitrogen fixation in root nodules. Science 2021 (DOI: 10.1126/science.abg2804).
D'Ario M, Tavares R, Schiessl K, Desvoyes B, Gutierrez C., Howard M and Sablowski R. Cell size controlled in plants using DNA content as an internal scale. Science 2021 (DOI: 10.1126/science.abb4348).
Yang W, Cortijo S, Korsbo N, Roszak P, Schiessl K, Gurzadyan A, Wightman R, Jönsson H, Meyerowitz E. Molecular mechanism of cytokinin-activated cell division in Arabidopsis. Science 2021 ( DOI: 10.1126/science.abe2305).
Schiessl K, Lilley J L S, Lee T, Tamvakis I, Kohlen W, Bailey P C, Thomas A, Luptak J, Ramakrishnan K, Carpenter M D, Mysore K S, Wen J, Ahnert S, Grieneisen V A and Oldroyd G E D: NODULE INCEPTION recruits the lateral root developmental program for symbiotic nodule organogenesis in Medicago truncatula. Current Biology 2019 (DOI: 10.1016/j.cub.2019.09.005).
Magne K, Couzigou J-M, Schiessl K., Liu S, George J, Zhukov V, Sahl L, Boyer F, Iantcheva A, Mysore K S, Wen J, Citerne S, Oldroyd G E D, Ratet P: MtNODULE ROOT1 and MtNODULE ROOT2 Are Essential for Indeterminate Nodule Identity. Plant Physiology 2018 (DOI: 10.1104/pp.18.00610).
Mislata A, Bencivenga S, Bush M, Schiessl K, Boden S and Sablowski R: DELLA genes restrict inflorescence meristem function independently of plant height. Nature Plants 2017 (DOI: 10.1038/s41477-017-0003-y).
Mislata A, Schiessl K, Sablowski R: Active control of cell size generates spatial detail during plant organogenesis. Current Biology 2015 (DOI:10.1016/j.cub.2015.10.008).
Schiessl K, Muino J M, Sablowski R: Arabidopsis JAGGED links floral organ patterning to tissue growth by repressing Kip-related cell cycle inhibitors. PNAS 2014 (DOI:10.1073/pnas.1320457111).
Sauret-Güeto S, Schiessl K, Bangham A, Sablowski R, Coen E: JAGGED Controls Arabidopsis Petal Growth and Shape by Interacting with a Divergent Polarity Field. PLOS Biology 2013 (DOI: 10.1371/journal.pbio.1001550).
Schiessl K, Kausika S, Southam P, Bush M, Sablowski R: JAGGED controls growth anisotropy and coordination between cell size and cell cycle during plant organogenesis. Current Biology 2012 (DOI: 10.1016/j.cub.2012.07.020).