Understanding the evolution of novelty through computational modelling
The most striking outcome of evolution is the emergence of novel forms of life, with new abilities and interesting shapes, colours and sizes.
In an Essays in Biochemistry review, Sainsbury Laboratory Cambridge University’s (SLCU) Enrico Sandro Colizzi and Renske M.A. Vroomans, together with Utrecht University’s Paulien Hogeweg, explore how computational models can give us new insights into how novelty evolves in biological systems.
Over millions of years, the evolution of novel traits has brought about complex cells from collections of molecules, and massive structures like trees and elephants from collections of cells.
Evolution builds on structures that already exist. For example, feathers were co-opted for flight after first being evolved for their heat insulation, sensory or display function. But then, how did a complex structure like a feather evolve at all?
It is sometimes difficult to imagine how a novel trait emerges as a consequence of random mutations in the DNA. For this reason, it is useful to construct mathematical models that allow scientists to simulate evolution and track how many small mutations combined lead to new features at the macroscale.
The paper’s first author, Dr Colizzi, Research Associate in the Locke Group at SLCU, explains that there is a catch when constructing such models: “If you incorporate the novel trait into the model, it isn’t really novel: the model already “knows” about the trait. But how do you study the evolution of a novel trait without building it into the model?”.
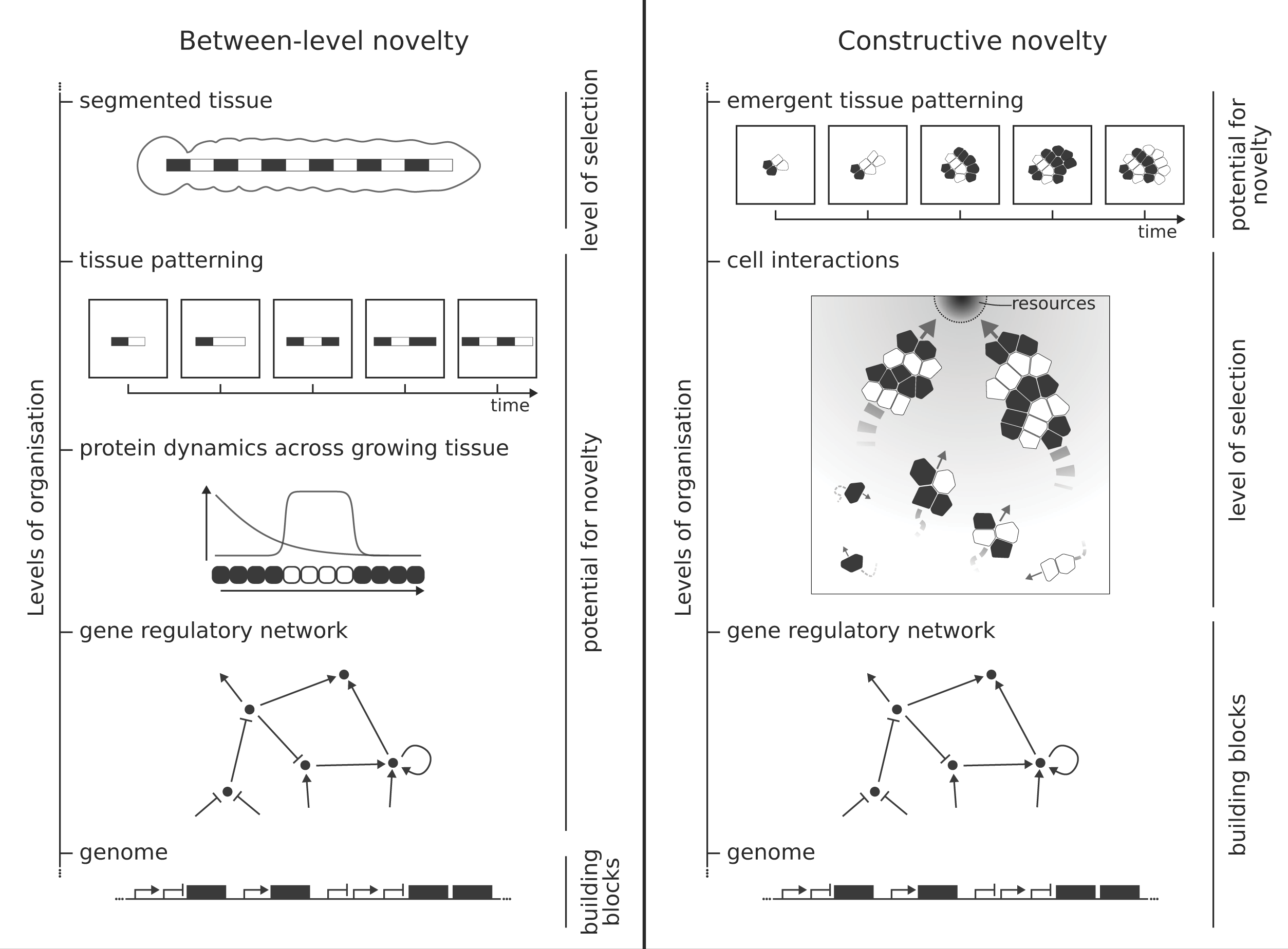
Between-level novelty and constructive novelty
“We identify two broad ways to construct models that do capture novelty, which we call ‘Between-level novelty’ and ‘Constructive novelty’, Dr Colizzi added. “In between-level models, you fix two levels of organisation, and let the mechanism evolve that generates the higher level (the whole organism) from the lower level (the genome).
“These models are useful for answering questions like: How do genetic mutations change embryonic development to generate a particular feature? How many different ways are there to generate this feature? For instance, vertebrates (animals with a spine, like us) and insects generate patterns of repeated body parts (e.g. vertebrae and abdomen segments) during embryonic development. In models that study how repeated segments evolved, the virtual animals that make more segments reproduce more frequently.
“In other words, segments are assumed to be possible and beneficial and therefore making segments is not a novelty. What is novel instead, is the way in which genes are used to specify how an individual embryo develops its segments.”
These models have shown that there are many different ways to make segments, some of which indeed match the ways vertebrates and insects make theirs. Scientists can use these models to better understand why vertebrates and insects develop the way they do.
Two forms of evolutionary novelty in computational models: Between-level novelty: Evolutionary novelty can arise as information transcoding from the genotype, the level where the model-building blocks are specified, to the phenotype, where selection takes place. Constructive novelty: Evolutionary novelty can arise as a side effect of selection on a tangentially related trait. In the case of an evolutionary transition, this can lead to a new level of organisation. The figure sketches two examples of models that capture these forms of novelty. Graphic originally published in Essays in Biochemistry by Portland Press Limited Portland Press Limited on behalf of the Biochemical Society and distributed under the Creative Commons Attribution License 4.0 (CC BY).
“In models of constructive novelty, we do not specify what the end-goal of evolution should be. Instead, in these models we give individuals something to do,” Colizzi said. “For example, we simulate cells that have to find food to survive, and we give these cells some tools that they can use to achieve this goal, for example they can decide when they divide and when they move around to search for food, by mutating their genetic information. These models can answer questions of the kind: What can evolve from some genetic building blocks to survive in a particular environment? For example, models of the evolution of multicellularity show how cells can evolve to stick to each other because groups of cells are better at finding food in complex environments. We think that these constructive models capture more closely how novel features evolve.”
While models are often constructed to study the emergence of a particular novel trait (segments, multicellularity), the authors expect that similarities will appear between different computer models as more of them are constructed. These similarities will hopefully point towards general principles of novelty evolution.
“We expect that the field of computational evolutionary developmental biology (evo–devo) will still reveal many more exciting mechanisms for the evolution of novelty,” added Dr Vroomans, who leads a research group at SLCU. “With ever-increasing computational resources, we hope to eventually construct computer models that really surprise us, where novelty evolves upon novelty, creating new ways to survive and thrive in the simulations that we had not even come up with when we started programming.”
Reference
Enrico Sandro Colizzi, Paulien Hogeweg, Renske M.A. Vroomans; Modelling the evolution of novelty: a review. Essays Biochem 8 December 2022; 66 (6): 727–735. doi: https://doi.org/10.1042/EBC20220069