Scientists often spend countless hours on tedious measurements. But one such measurement just got a lot easier - counting fungal structures to quantify arbuscular mycorrhizal fungi (AMF) colonisation in plant roots is now automated thanks to a new method developed at the Sainsbury Laboratory Cambridge University (SLCU).
Understanding how soil fungi establish beneficial relationships with the roots of plants and how this relationship could be enhanced is of growing importance as we look for more sustainable methods to increase plant nutrition and yield.
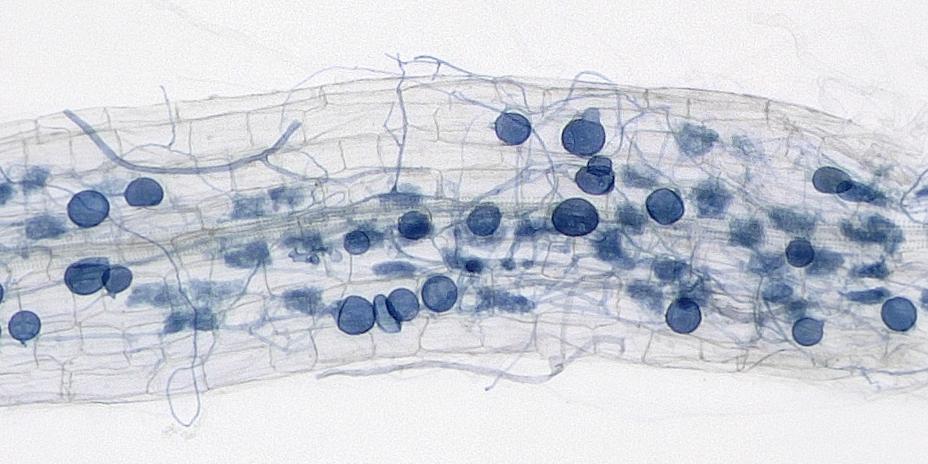
Quantifying the proportion of roots colonised by AMF is a routine task, but it is a slow, manual task and susceptible to human error.
One of the most commonly used, and considered to be the most reliable, method was developed decades ago and involves undertaking a visual assessment of the root samples. Called the grid-line intersect method, it uses traditional microscopy techniques to estimate the proportion of infected roots. This method involves the scientist drawing a grid on a petri dish, staining the fungal structures and then peering down a low-resolution dissecting microscope and using a handheld click counter to do the assessment.
It was while visiting SLCU as an undergraduate and doing some AMF counting herself, that Alice McDowell, now undertaking her PhD in Biochemistry at Cambridge, came up with the idea of an automated system that identifies and then quantifies fungal structures.
Alice’s idea was taken up by Carl Turner, from Cambridge’s Department of Applied Mathematics and Theoretical Physics. Dr Turner designed the first prototype of the software. And Sebastian Schornack’s team from SLCU pursued software development and expanded its capabilities. The result is an automatic computer vision-based system to identify and quantify AM fungal colonisation called the Automatic Mycorrhiza Finder (AMFinder).
“It is a highly reliable way to document and quantify fungal structures inside segments of plant roots and even whole plant root systems,” said Edouard Evangelisti, who led the AMFinder’s development and is the first author of the research published in New Phytologist. “We have developed and tested it on multiple plant hosts, it can identify different types of fungal structures in roots thanks to colleagues providing a rich resource of colonised plant root images. We have also been able to measure the dynamics of colonisation over time. It is still a simple process to prepare the samples with a straightforward staining protocol and image them with a flatbed scanner or digital microscope.”
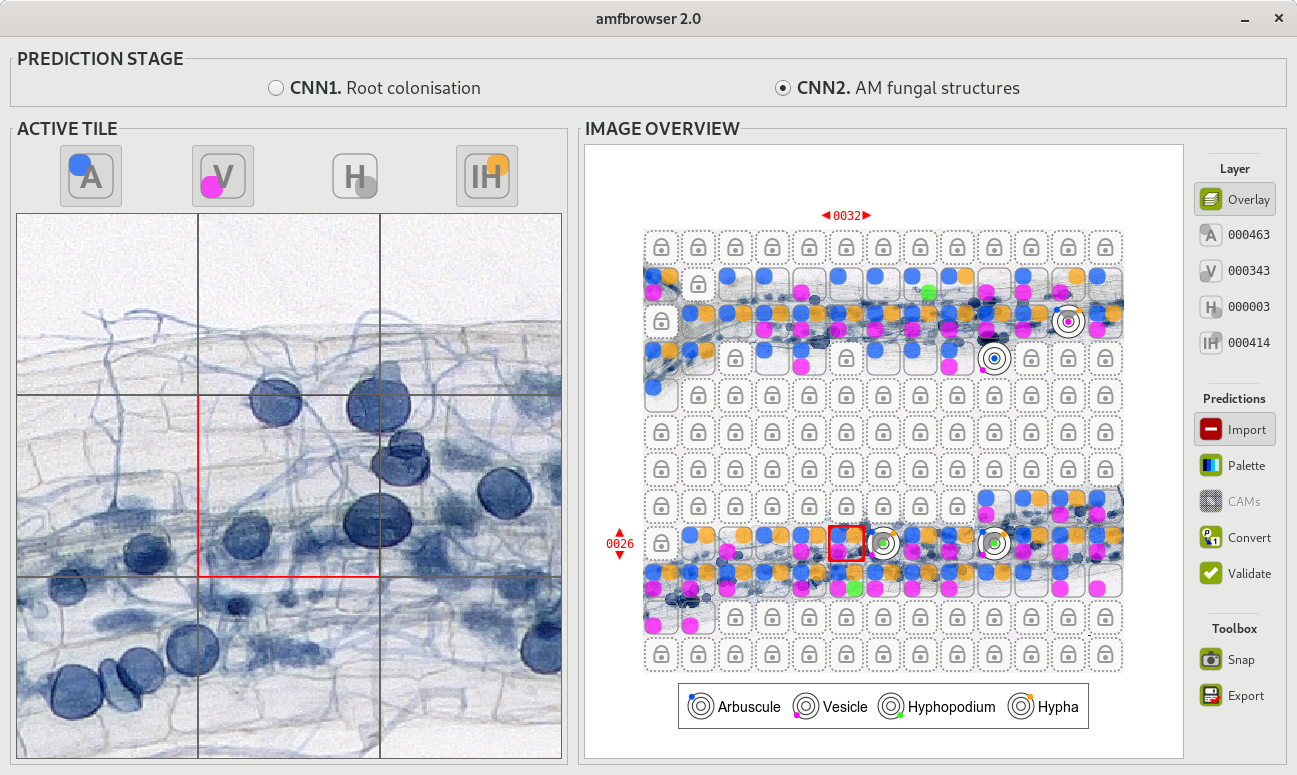
AMFinder interface: AMFinder is used either to predict fungal colonisation and intraradical hyphal structures within plant root images (prediction mode), or to train AMFinder neural networks (training mode).
AMFinder was developed using a deep learning system called convolutional neural networks (CNNs). This is a class of artificial neural network is often used to analyse images, where the computer model learns to extract relevant information from pre-analysed data. AMFinder adapts to a wide array of experimental conditions and produces accurate and reproducible analyses of plant root systems. Not only that, “AMFinder improves the way researchers document root colonisation by allowing them to go back to archived computer annotations if needed,” added Sebastian Schornack.
AMFinder accurately identifies colonised root sections and intraradical hyphal structures in several plant species commonly used in mycorrhiza research, including Nicotiana benthamiana, Medicago truncatula, Lotus japonicus, and Oryza sativa, and is compatible with the AM fungi Rhizophagus regularis, Claroideoglomus claroideum, Rhizoglomus microaggregatum, Funneliformis geosporum and Funneliformis mosseae.
This article is also highlighted in FacultyOpinions.
AMFinder Tool
AMFinder can be accessed at https://github.com/SchornacklabSLCU/amfinder.
Reference
Edouard Evangelisti, Carl Turner, Alice McDowell, Liron Shenhav, Temur Yunusov, Aleksandr Gavrin, Emily K. Servante, Clément Quan, Sebastian Schornack (2021) Deep learning-based quantification of arbuscular mycorrhizal fungi in plant roots, New Phytologist