Research Interests
My research is centred on investigating the infectious strategies employed by symbiotic microorganisms during their beneficial interactions with plants. Specifically, I have a keen interest in exploring the molecular functions of proteins secreted by these microorganisms, known as "effectors," and their impact on plant immunity, physiology, and development.
In the context of plant-microbe interactions, the secretion of effector proteins plays a pivotal role in the ability of pathogenic microorganisms to bypass the plant's immune system and establish infection. However, despite extensive research on pathogenic effectors, our understanding of effectors secreted by symbiotic microorganisms and their functions during beneficial interactions remains limited. Through my research, I aim to advance our knowledge of these protein classes and shed light on the adaptive mechanisms governing plant physiology during beneficial interactions.
Research background
My academic background lies in plant molecular biology and the study of plant-microbe interactions. After completing my final internship in the laboratory of Eric Giraud (IRD, Montpellier, France), I was awarded a doctoral grant to continue my research. During my doctoral studies, I focused on the molecular characterization of effectors secreted by symbiotic bacteria through the T3 secretion system during nitrogen fixation symbiosis. Specifically, my research involved identifying and characterizing a novel class of rhizobial effectors capable of inducing the development of nitrogen-fixing nodules in legume plants even in the absence of Nod-factors. Following the completion of my Ph.D. in 2019, I continued my research as a Post-Doc/Research Associate in the same laboratory for an additional year.
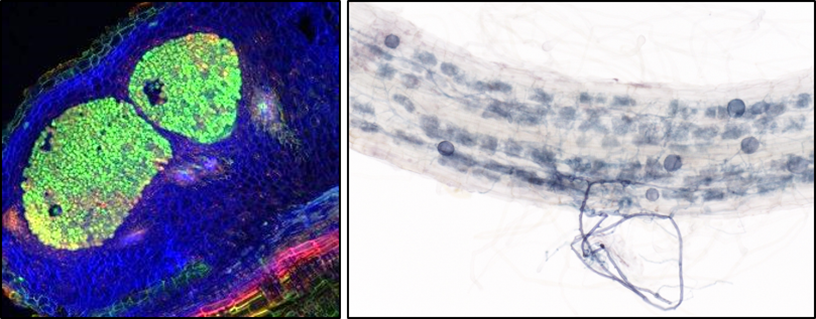
Two types of close intimate symbiotic interactions. On the left, symbiotic bacteria (green) colonize the cells of newly formed root organs, providing nitrogen to the plant. (Image by Albin Teulet). On the right, the symbiotic fungus (blue) forms arbuscules inside plant root cells, facilitating a mutual exchange of nutrients that benefits both the fungus and the plant (Image by Edwige Berthelot)
Research in the Schornack Group
In 2021, I made the decision to expand the scope of my research to encompass other symbiotic interactions and joined the Schornack Group at SLCU. My current projects involve the molecular characterization of the newly discovered MycFOLD effector family secreted by arbuscular mycorrhizal fungi, focusing on its role in the arbuscular mycorrhizal (AM) symbiosis. Additionally, I am investigating the involvement of immune signaling pathways in the establishment of the AM symbiosis from the perspective of the plant.
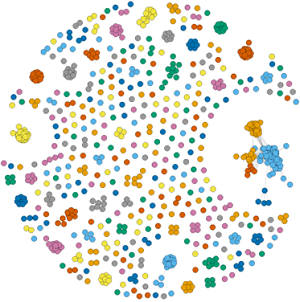
Structural clustering of predicted effector proteins from the symbiotic fungus Rhizophagus irregularis (From Teulet et al., 2023)
Through these projects, I aim to deepen our understanding of the intricate relationships between microorganisms and their hosts and identify potential targets to enhance the efficacy of beneficial microorganisms in various applications.
Key publications
Teulet A, Quan C, Evangelisti E, Wanke A, Yang W, Schornack S. A Pathogen Effector FOLD Diversified in Symbiotic Fungi. In press in New Phytologist 2023 Twitter thread summary [https://twitter.com/dromius/status/1604054543557328896?s=20]
Teulet A*, Camuel A*, Perret X*, Giraud E*. The Versatile Roles of Type III Secretion Systems in Rhizobium-Legume Symbioses. Annu Rev Microbiol. 2022 76: 211-233 - doi: 10.1146/annurev-micro-041020-032624
Busset N, Gully D, Teulet A, Fardoux J, Cornu D, Barrachina C, Giraud E, Mergaert P. The type III effectome of the symbiotic strain ORS3257. Biomolecules 2021 11: 1592 - doi: 10.3390/biom11111592.
Ratu STN, Teulet A, Miwa H, Masuda S, Nguyen HP, Yasuda M, Sato S, Kanek T, Hakoyama T, Hayashi M, Giraud E, Okazaki S. Rhizobium use a pathogenic-like effector to hijack leguminous nodulation signalling. Scientific Report 2021 11:2034 – doi: 10.1038/s41598-021-81598-6
Teulet A, Gully D, Rouy Z, Koebnik R, Giraud E, Lassalle F. Phylogenetic distribution and evolutionary dynamics of nod and T3SS genes in genus Bradyrhizobium. Microbial genomic 2020 6(9): mgen000407. doi: 10.1099/mgen.0.000407
Teulet A, Busset N, Fardoux J, Gully D, Chaintreuil C, Cartieaux F, Jauneau A, Comorge V, Okazaki S, Kaneko T, Gressent F, Nouwen N, Arrighi JF, Koebnik R, Mergaert P, Deslandes L, Giraud E. The rhizobial type III effector ErnA confers the ability to form nodules in legumes. Proc Natl Acad Sci USA 2019 116(43): 21758-21768. doi: 10.1073/pnas.19044561.
Okazaki S, Tittabutr P, Teulet A, Thouin J, Fardoux J, Chaintreuil C, Gully D, Arrighi JF, Furuta N, Miwa H, Yasuda M, Nouwen N, Teaumroong N, Giraud E. Rhizobium legume symbiosis in the absence of Nod factors two possible scenarios with or without the T3SS. ISME J 2016 10: 64-74. doi: 10.1038/ismej.2015.103.
* authors contributed equally to this study