Research interests
My overarching research interest lies in understanding the dynamic mechanisms and developing the tools that reveal how plants adapt and develop. During my PhD I focused on plant-pathogen interactions and virus transmission, I am now exploring plant developmental biology. Throughout my career, a recurring focus of my work has been the way plants integrate information, whether in the context of defence or development. My transition from plant-pathogen interactions to developmental biology allows me to build on the molecular approaches I developed during my PhD, where cell-to-cell communication and molecular signalling already played a central role. I am now particularly interested in how these regulatory signals shape pattern formation and contribute to developmental plasticity.
Research background
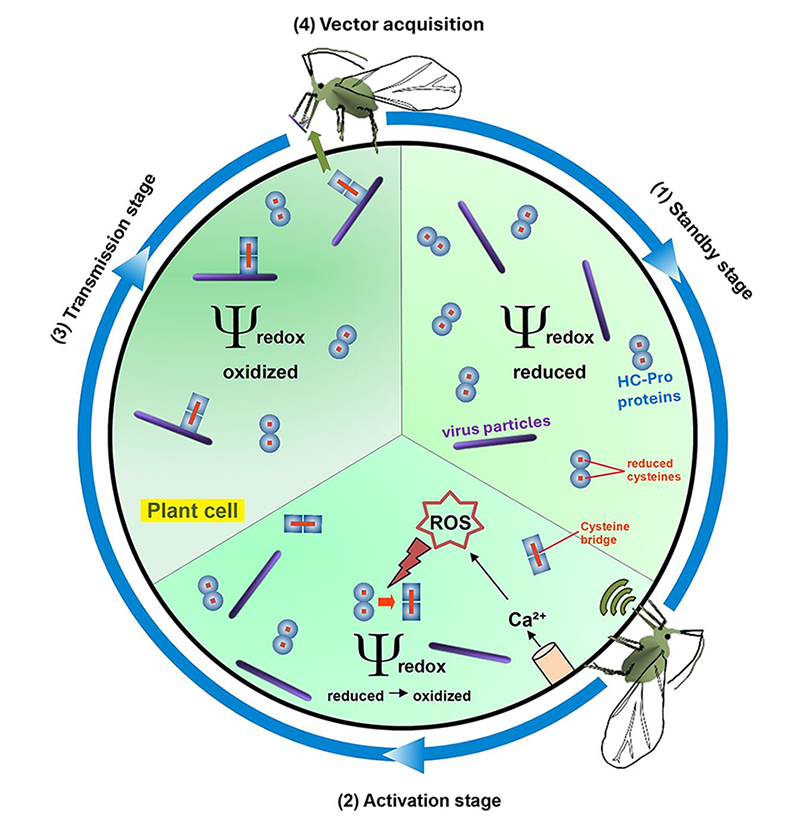
I specialised in plant-microbe molecular interactions during my undergraduate studies in Montpellier (France), which led me to pursue a PhD focused on virus transmission in plants. I carried out my doctoral research in the group of Dr. Drucker, in collaboration with the FN3PT (Fédération Nationale des Producteurs de Plants de Pomme de Terre). My project aimed to understand how Turnip mosaic virus (TuMV) is transmitted to plants by aphids, with a particular focus on the viral protein HC-Pro (Helper Component Protease), which plays a key role in facilitating virus transmission. I discovered that when aphids feed on TuMV-infected plants, they trigger calcium signalling and the production of reactive oxygen species (ROS) in the host tissues. These signals alter the redox state of the plant cells and promote the formation of disulfide bridges between HC-Pro monomers, leading to the appearance of protein dimers. Strikingly, the accumulation of HC-Pro dimers correlates with an increased rate of virus transmission (Berthelot et al., 2019).
Aphid feeding triggers a redox-dependent switch during TuMV transmission.
Although my PhD was rooted in plant-pathogen interactions, this work revealed how plants perceive and respond to biotic stimuli by modulating their internal cellular environment. These responses illustrate the capacity of plant cells to dynamically integrate external cues, a concept that resonates with the broader theme of developmental plasticity.
Current Research in the Moyroud Group
I joined the Moyroud group in 2023 and my current research focuses on understanding how cellular and molecular mechanisms establish petal patterning in Hibiscus trionum. This model system provides a fascinating opportunity to investigate the formation of the "bullseye" pattern in petals, where distinct regions of the petal surface are characterized by differences in pigmentation, morphology, and cuticle structure of epidermis cells. Specifically, I am interested in identifying the genes responsible for these cellular differences, particularly those involved in the specification of the bullseye boundary between the proximal (pigmented) and distal (non-pigmented) regions of the petal.
Using RNA sequencing, we have identified several genes that are preferentially expressed in different petal regions, and I am investigating their roles in petal development. My work involves characterizing cellular morphology during key stages of bullseye formation, generating reporter lines to visualize gene expression, and creating CRISPR lines to investigate boundary gene function. I am also interested in whether genes that pattern the petal surface also control cell differentiation, and particularly the specification of mucilage cells, in the internal petal tissues. Additionally, I am developing an in situ hybridization protocol tailored to Hibiscus to localize gene expression within specific petal tissues. Through this work, we aim to uncover how robust boundaries are established and maintained during petal growth, shedding light on the broader mechanisms of plant pattern formation.
Petal anatomy along the proximo-distal axis during early stages of bullseye formation in Hibiscus trionum.
Cell type diversity and mucilage cell maturation in Hibiscus trionum petals.
Work in Progress
- Designing reporter lines using mTurquoise to visualize boundary-specific gene expression in Hibiscus trionum.
- Generating CRISPR lines to target genes expressed at the bullseye boundary.
- Characterising mucilage cell development and the mechanisms underpinning their specification.
- Developing an in situ hybridization protocol to study gene expression in Hibiscus petals.
Science communications and outreach
I care deeply about creating inclusive and welcoming spaces where people can connect, share ideas, and enjoy meaningful moments together. This motivation has led me to be actively involved in organising both scientific and outreach events throughout my career.
During my PhD, I served for three years on the organising committee for the UMR Biologie et Génétique des Interactions Plante-Parasite weekly seminar series, including one year as chair, and I coordinated the Printemps de Baillarguet, a two-day campus-wide event highlighting the work of non-permanent employees. I also created visual templates and organisational guides that helped sustain these initiatives and inspired similar events on other campuses.
At the Sainsbury Laboratory, I continue to foster public engagement with science. I have developed interactive science activities, including a plant maze to demonstrate root and shoot growth dynamics and response to environment, and regularly volunteer at outreach events in Cambridge such as the Festival of Plants and Big Biology Day. Most recently, I worked on an award-winning interactive plant science exhibit for the RHS Chelsea Flower Show, helping with preparation, set-up and talking with visitors about the research our Laboratory does.
Training and development of the next generation
I have also supported science education through student training programmes, including mentoring a T-level student for six months and supervising students in the Aspiring Scientists Training Programme for the past two years. I am currently part of the organising team for the Sainsbury Laboratory’s 2025 Retreat.
Key publications
Berthelot E*, Macia JL, Martinière A, Morisset A, Gallet R, Blanc S, Khelifa M, Drucker M (2019). Pharmacological analysis of transmission activation of two plant viruses, Turnip mosaic virus and Cauliflower mosaic virus Scientific Reports 9:9374. https://doi.org/10.1038/s41598-019-45904-7
Berthelot E*, Ducousso M, Macia JL, Baecker V, Thébaud G, Gallet R, Yvon M, Blanc S, Khelifa M, Drucker M (2019). Turnip mosaic virus is a second example of a virus using transmission activation for plant-to-plant propagation by aphids. Journal of Virology e01822-18. https://doi.org/10.1128/jvi.01822-18
Dáder B*, Then C*, Berthelot E*, Ducousso M, Ng JCK, Drucker M (2017). Insect transmission of plant viruses: Multilayered interactions optimize viral propagation. Insect Science 24:929-946. https://doi.org/10.1111/1744-7917.12470