Dr Sebastian Ahnert was a Career Development Fellow at SLCU between 2016-2020. This page summarises the research areas that his former group focused on.
Ahnert Group
The focus of our work lies on the interface of theoretical physics, biology, mathematics and computer science. We are particularly interested in using algorithmic descriptions of structures and functional systems in order to quantify and classify their complexity. Examples of the application of such approaches include pattern detection in gene expression data, the classification of protein quaternary structure (see below), the structure of genotype-phenotype maps (see below) and the identification of large-scale features in complex networks. Connected to this is also an interest in interdiscliplinary applications of network analysis, particularly in the context of digital methods and large-scale data analysis in the humanities.
Protein Quaternary Structure
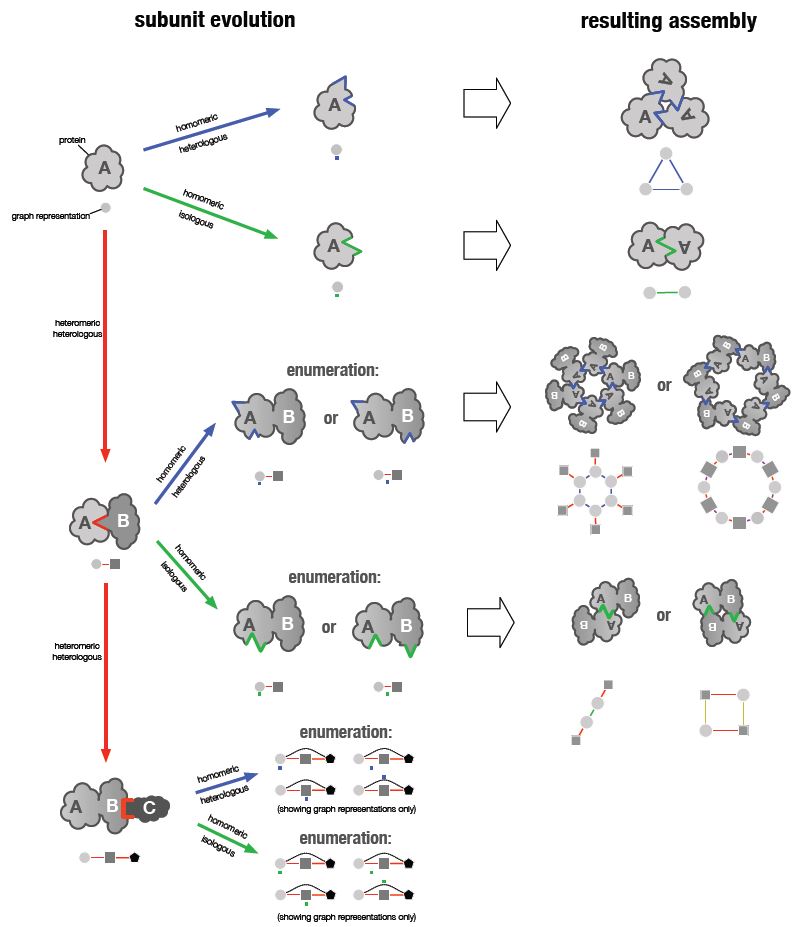
Biological evolution has produced an enormous variety of protein complexes, which arise when several proteins bind together to form larger structures. In recent work we have shown that the vast majority of protein complexes can be broken down in terms of three different fundamental steps of protein evolution. These steps can combine in many different ways, giving rise to the observed variety of protein complexes. What this reveals is that heteromeric protein complexes, which are complexes that consist of more than one type of protein, can be represented as homomeric complexes of repeated multi-protein units. This approach also allows us to classify protein complexes in a periodic table, and to predict topologies of complexes that have not been observed yet.
Genotype-Phenotype Maps
We are also interested in general properties of genotype-phenotype (GP) maps, and the effect these properties have on evolutionary outcomes. Such properties include a highly skewed distribution of genotypes per phenotype, strong correlations between the phenotypes of neighbouring genotypes in the network of point mutations, and a positive correlation between phenotypic evolvability and robustness. We can study these properties using the RNA secondary structure GP map, the HP model of protein folding, and a new GP map, the Polyomino GP map, which can be viewed as a model of protein quaternary structure self-assembly. The Polyomino map can also be used to study the evolution of phenotypic complexity and the properties of non-deterministic GP maps.
Key publications
S. E. Ahnert, Structural properties of genotype-phenotype maps,
Journal of the Royal Society Interface 14, 20170275 (2017)
S. E. Ahnert, J. A. Marsh, H. Hernandez, C. V. Robinson, S. A. Teichmann
Principles of assembly reveal a periodic table of protein complexes
Science 350, 1331 (2015)
J. Marsh, H. Rees, S. E. Ahnert, S. A. Teichmann
Structural and evolutionary versatility in protein complexes with uneven stoichiometry Nature Communications 6, 6394 (2015)
M. Taylor-Teeples, L. Lin, M. de Lucas M, G. Turco, T. W. Toal, A. Gaudinier, N. F. Young, G. M. Trabucco, M. T. Veling, R. Lamothe, P. P. Handakumbura, G. Xiong, C. Wang, J. Corwin, N. Tsoukalas, L. Zhang, D. Ware, M. Pauly, D. J. Kliebenstein, K. Dehesh, I. Tagkopoulos, G. Breton, J. Pruneda-Paz, S. E. Ahnert, S. A. Kay, S. P. Hazen, S. M. Brady
An Arabidopsis Gene Regulatory Network for Xylem Specification and Secondary Wall Biosynthesis Nature 517, 571 (2015)
S. F. Greenbury, I. G. Johnston, A. A. Louis, S. E. Ahnert A tractable genotype-phenotype map modelling the self-assembly of protein quaternary structure Journal of The Royal Society Interface 11, 20140249 (2014)
J. A. Marsh, H. Hernandez, Z. Hall, S. E. Ahnert, T. Perica, C. V. Robinson, S. A. Teichmann Protein complexes are under evolutionary selection to assemble via ordered pathways Cell 153, 461 (2013)
M. A. Moreno-Risueno, J. M. Van Norman, A. Moreno, J. Zhang, S. E. Ahnert, P. N. Benfey Oscillating Gene Expression Determines Competence for Periodic Arabidopsis Root Branching Science 329, 1306 (2010)
S. E. Ahnert, I. G. Johnston, T. M. A. Fink, J. P. K. Doye, A. A. Louis Self-assembly, modularity and physical complexity Physical Review E 82, 026117 (2010)
S. E. Ahnert, K. Willbrand, F. C. S. Brown, T. M. A. Fink Unbiased pattern detection in microarray data series Bioinformatics 22, 1471 (2006)